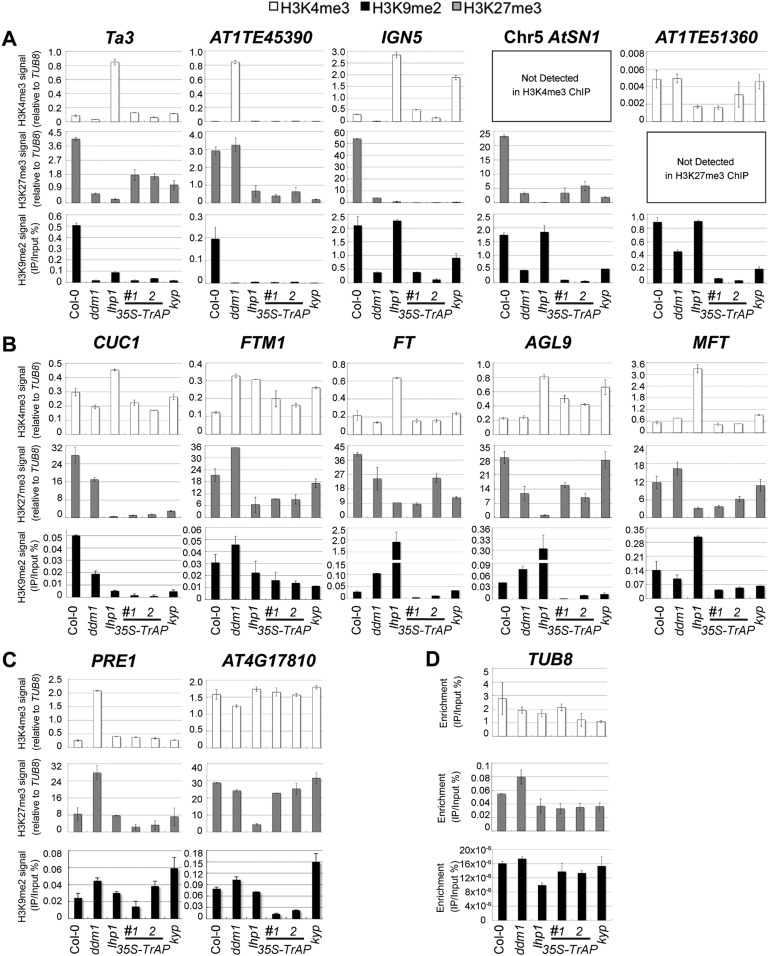
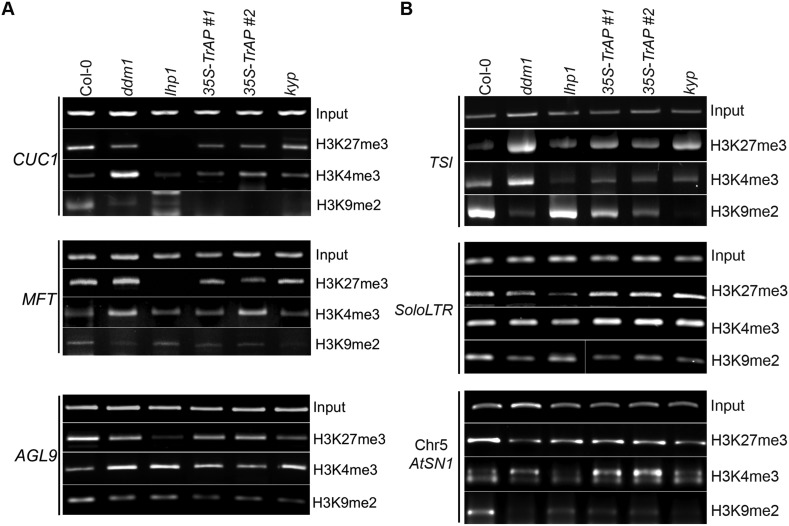
Figure 6. ChIP-qPCR analyses of H3K4me3, H3K9me2, and H3K27me3 in TrAP-regulated loci in vivo.
(A) TrAP-activated transposons in heterochromatic regions contained reduced H3K9me2 and H3K27me3 but did not show consistent variation in H3K4me3 marks. (B) TrAP-upregulated flowering genes showed consistently reduced H3K9me2 and H3K27me3 marks compared to wild-type Col-0. (C) TrAP-downregulated genes displayed variable changes of H3K9me2 and H3K27me3 marks and no obvious changes of H3K4me3 mark. (D) Tubulin (TUB8) was used as internal control for all the ChIP experiments; the percentage enrichment vs input is shown. ChIP assays were conducted on 11-day-old seedlings using antibodies specific for H3K9me2 (Abcam, Cat# ab1220), H3K27me3 (Millipore, Cat# 07-449), and H3K4me3 (Millipore, Cat# 04-745). Enrichment of H3K4me3 and H3K27me3 in each locus is normalized to that of TUB8; H3K9me2 enrichment is plotted as percentage of input. The standard deviation (SD) was calculated from at least three biological repeats.