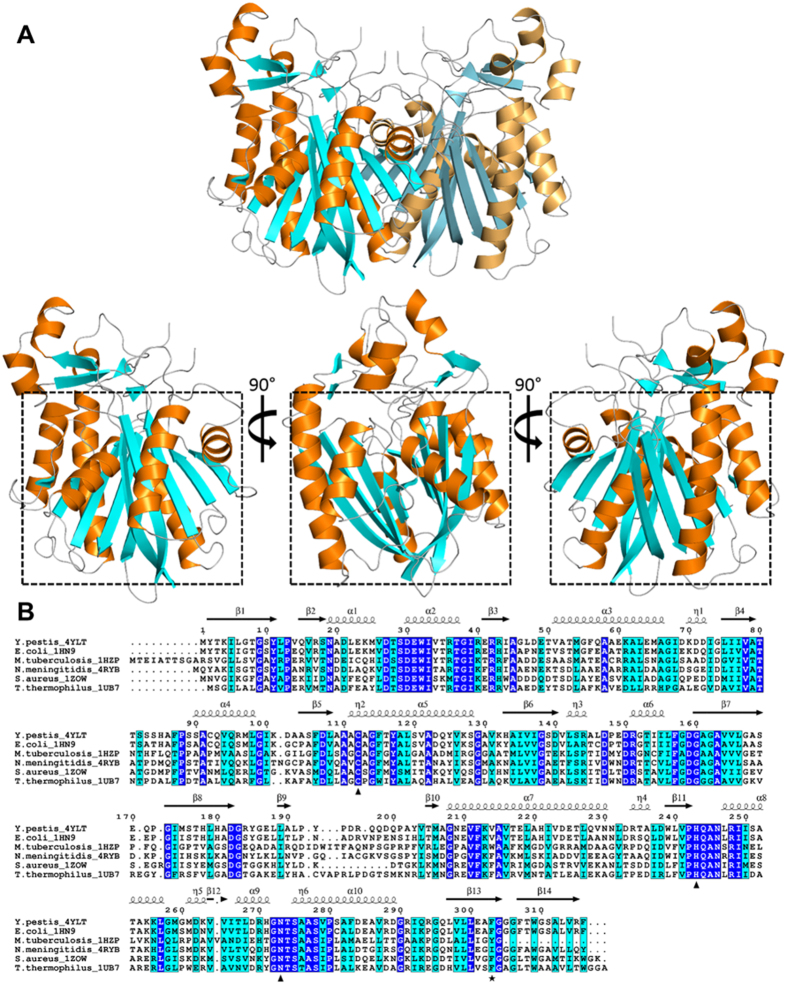
Figure 1. The structure of Yersinia pestis FabH (YpFabH).
(A) A cartoon representation of YpFabH as a dimer, and a monomer at 0°, 90°, and 180° of rotation around the horizontal axis, with the thiolase fold motif highlighted by a black square. β-sheets are displayed in cyan, α-helices are displayed in orange. (B) Sequence alignment of YpFabH with FabH from E. coli (PDB:1HN9), M. tuberculosis (PDB:1HZP), N. meningitidis (PDB:4QAV), S. aureus (PDB:2GQD), and T. thermophilus (PDB:1J3N). Strictly conserved residues are highlighted in blue with white text, similar residues are highlighted in cyan, residues of the active site catalytic triad are designated by triangles, residue Phe303, which is thought to play a role in substrate specificity, is designated by a black star. Secondary structure features of YpFabH, including α-helices, β-sheets, and 3-10 helices (η) are shown above the sequence alignment in black.