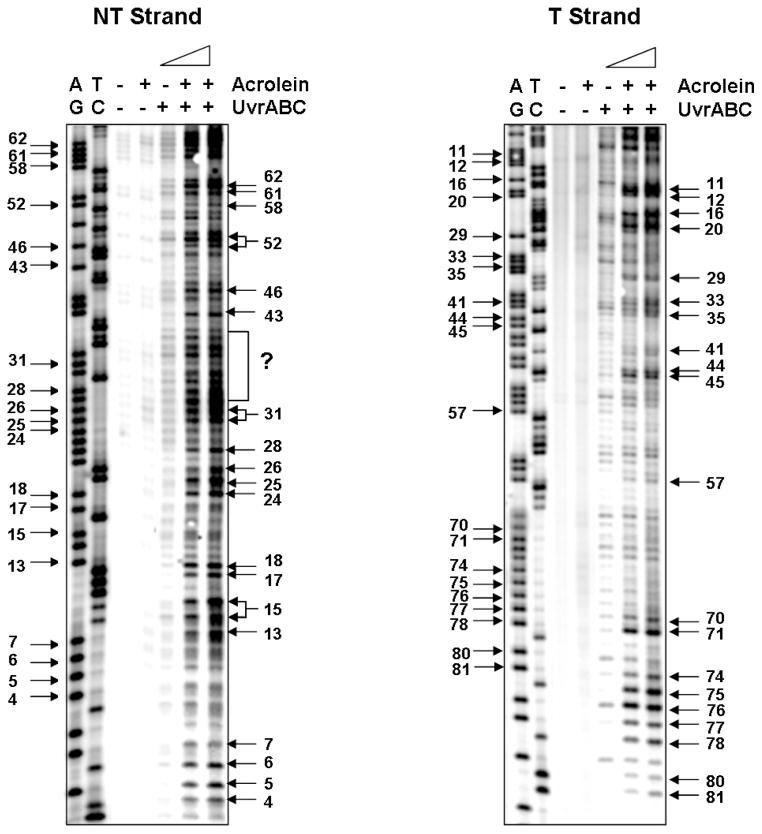
Figure 6.
The Acr-dG adduct distribution detected by UvrABC incision method in the supF gene sequence. The supF DNA fragments were prepared from pSP189 plasmid DNA modified with different concentrations of Acr, single 5′-end labeled with 32P at the nontranscribed stand (NT strand) or at the transcribed strand (T strand), reacted with UvrABC, and the resultant DNAs were separated by 8 % denatured polyacrylamide gel electrophoresis as described in Materials and Methods. A) represents a typical radiogram resulting from single 5′-end labeled with 32P at the transcribed strand, and B) represents a typical radiogram resulting from single 5′-end labeled with 32P at the nontranscribed strand. Symbols: GA and TC represent Maxam and Gilbert sequencing reaction products, and +/− represents reaction with Acr or UvrABC nuclease. Guanine residue numbers in the sequence are labeled at the left and the corresponding UvrABC incision bands are labeled at the right. Note: we were unable to assign the UvrABC incision bands within the (?) area to corresponding guanine residues in the supF sequence.