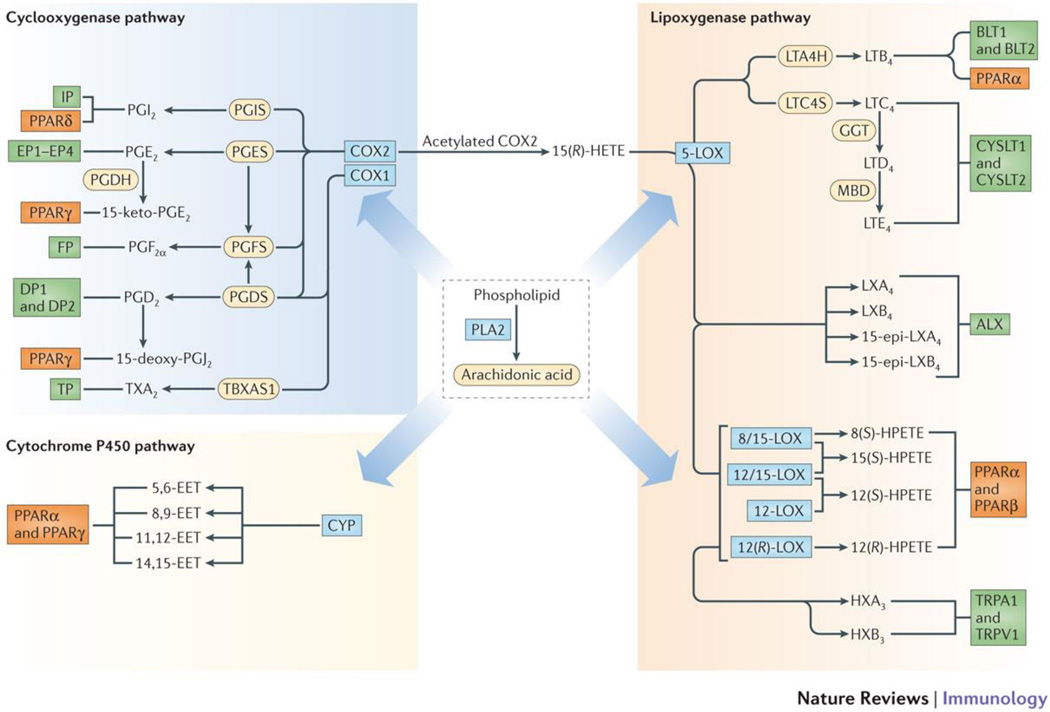
Figure 1. Eicosanoid biosynthesis and receptor signaling.
Lipidomic view of phospholipase A2 (PLA2), cyclooxygenase-1 and -2 (COX1/2), 5-lipoxygenase (5-LOX), 8-, 12-, and 15-lipoxygenases (8/12/15-LOX) and cytochrome P450 epoxyhydrolase (CYP) pathways of eicosanoid biosynthesis derived from arachidonic acid. Downstream enzymes are shown as yellow ovals followed by eicosanoid species and their receptors as green boxes. The peroxisome-proliferator activating receptors (PPARs), indicated as orange boxes, that are potentially activated by the eicosanoids are also shown.. The CYP ω-hydroxylase pathway and lipid species derived from other fatty acids are not shown. Abbreviations: COX, cyclooxygenase; CYP, cytochrome P450; EET, epoxyeicosatrienoic acid; GGT, gamma-glutamyl transferase; HETE, hydroxyeicosatetraenoic acid; HX, hepoxilin; LOX, lipoxygenase; LT, leukotriene; LTAH, LTA4 hydrolase; LTCS, LTC4 synthase; LX, lipoxin; MBD, membrane bound dipeptidase; PLA2, phospholipase A2; PPAR, peroxisome proliferator-activated receptor; TRPA1, transient receptor potential ankyrin 1; TRPV1, transient receptor potential vanilloid 1; and TX, thromboxane.