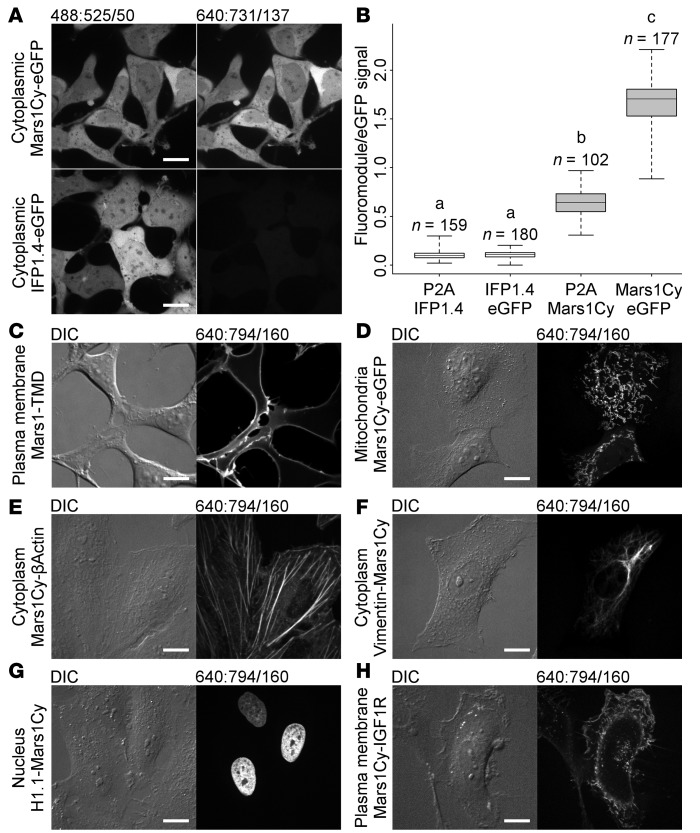
Figure 2. Fluoromodules appear brighter than IFP1.4 and are functional when expressed within mammalian cells.
Channels are labeled DIC or formatted excitation:emission/bandwidth. (A) HEK293 cells expressing Mars1Cy-eGFP or IFP1.4-eGFP fusion proteins, from otherwise identical vectors, were imaged in the presence of 400 nM SC1 and 50 μM biliverdin, respectively, to compare observed brightness under identical conditions and display settings. Images are representative samples of data analyzed in B. (B) Box plot of fluoromodule/eGFP signal intensity ratios measured from protein coexpressed with eGFP in HEK293 cells as fusion proteins or separate polypeptides (P2A). Whiskers indicate maximum and minimum values; for boxes, top, line, and bottom indicate first, second, and third quartiles, respectively. Differences between mean ratios were analyzed by 1-way ANOVA: F(3,614) = 5332.14, P < 0.001. A Games-Howell post-hoc test shows differences between paired group means as denoted by lowercase letters: a (IFP1.4-eGFP fusion versus separate IFP1.4/eGFP peptides, P = 0.650); other pairs (ab, ac, bc) describe different means with high confidence, P < 0.001. Number of measurements (n) per group (IFP1.4/eGFP: 159, IFP1.4-eGFP: 180, Mars1Cy/eGFP: 102, Mars1Cy-eGFP: 177) indicate the number of unique cells measured. (C) Mars1Cy anchored to the plasma membrane by a CD80 transmembrane domain (TMD) can be labeled with membrane-impermeant SCi1. (D) Signal peptide–prefixed (COXIV) Mars1Cy-eGFP is directed to the mitochondria and becomes labeled upon addition of the membrane-permeant fluorogen SC1. (E–H) Mars1- and Mars1Cy-based fluoromodules can be used to tag proteins such as β-actin, vimentin, histone H1.1, and IGF1R in living cells. Images in C–H represent at least 10 fields of view per transgene-hosting cell population. Scale bars: 16 μm.