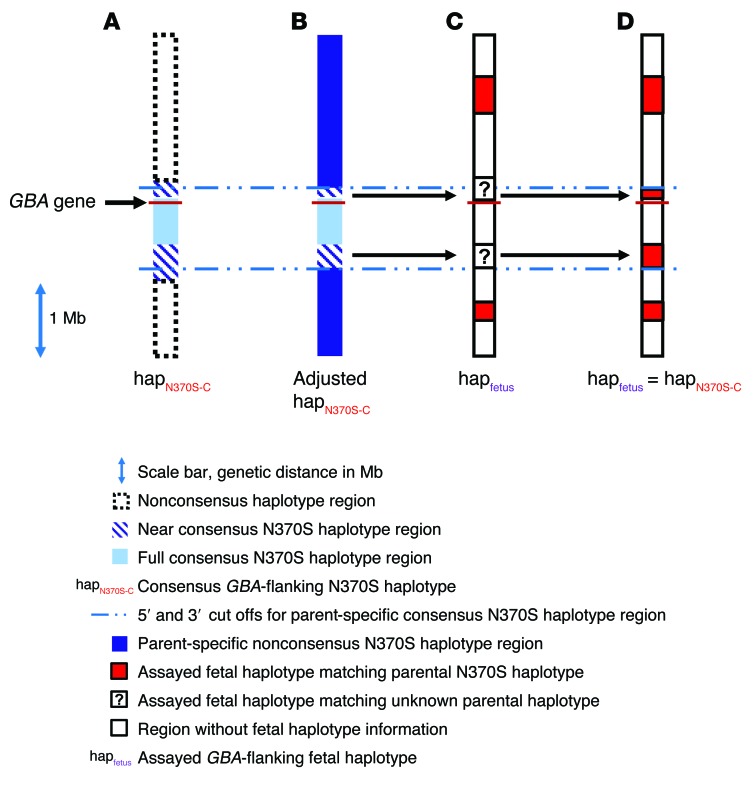
Figure 4. Extended fine mapping of the consensus and near-consensus AJ N370S founder haplotype region as a tool for phasing fetal haplotypes.
Illustration depicting the GBA locus (±2 Mb) and thousands of SNPs that were deep sequenced for the construction and typing of fetal alleles according to the analytical pipeline (as indicated in the key). (A) An extended deep-sequencing panel was used to better fine map the conserved N370S founder haplotype, as in Figure 2. Accordingly, a 301-SNP haplotype (termed “full-consensus N370S haplotype”) was identified in all N370S chromosomes in this study (28 chromosomes altogether). In addition, the consensus haplotype was found to extend 500 kb further downstream of GBA (620 additional SNPs) in 15 of 16 chromosomes from 8 N370S homozygotes. Furthermore, in all N370S homozygotes (but not all N370S carriers), the consensus haplotype was found to extend another 120 kb upstream of GBA (100 additional SNPs). Altogether, these extended haplotypes were termed “near-consensus N370S haplotypes.” (B) The N370S haplotype from each N370S carrier parent in the study was carefully mapped according to homozygous regions and family-based linkage analysis. After comparison to the near-consensus haplotype in A, new parent-specific 5′ and/or 3′ demarcations of the N370S near-consensus haplotype were set (this haplotype was termed the “parent-specific consensus N370S haplotype”). (C) In this example, deep sequencing of the GBA-flanking region in a fetus identified stretches of a linkage-based parental N370S haplotype that resided outside of the consensus N370S region. In addition, some stretches of fetal sequence could not be phased according to family-based linkage. (D) When unphased fetal sequence, such as in C, fell within the parent-specific consensus N370S haplotype (as determined in B), the consensus information was used to phase the fetus (here, with the N370S-linked haplotype), thereby increasing confidence in the diagnostic test result.