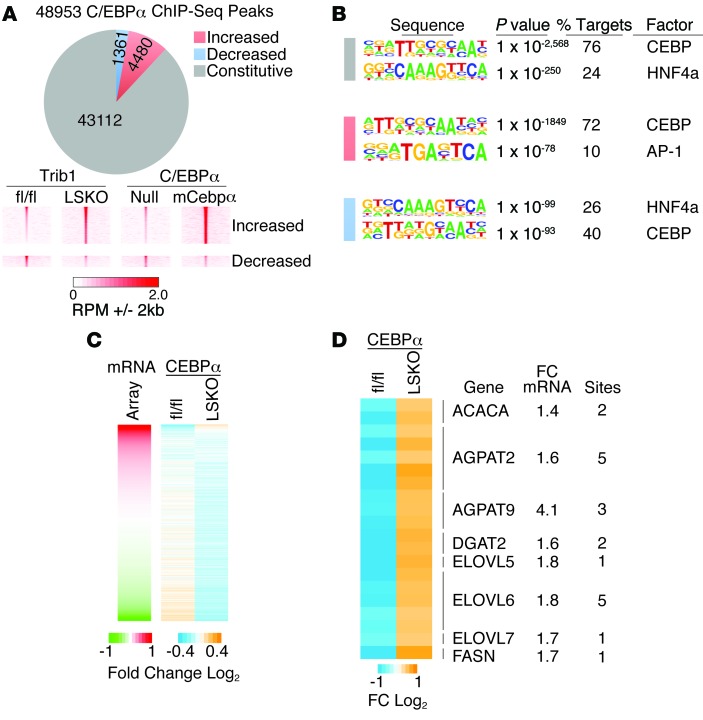
Figure 7. Trib1_LSKO mice have increased C/EBPα occupancy near lipogenic genes.
(A) Pie chart of all C/EBPα binding sites identified in liver by ChIP-seq showing the number of sites that were unchanged (constitutive), increased or decreased in Trib1_LSKO mice compared with Trib1_fl/fl mice. Density heat maps (lower panel) of raw sequence tags at regions differentially occupied by C/EBPα in Trib1_LSKO mice are shown for Trib1_fl/fl, Trib1_LSKO, AAV_Null, and AAV_mCebpa mice. (B) HOMER de novo motif analysis of constitutive and differential C/EBPα-binding sites in Trib1_LSKO mice. (C) Relationship of gene expression to C/EBPα occupancy. Left panel shows the relative expression of all genes in the microarray ordered from highest to lowest fold-change in Trib1_LSKO vs. fl/fl control. Right heat map depicts C/EBPα occupancy in Trib1_fl/fl and Trib1_LSKO mice at sites within 100 kb of the TSS. (D) Heat map of differential C/EBPα binding sites within 100 kb of the TSS for genes involved in hepatic lipogenesis. The number of C/EBPα sites and the microarray fold change are indicated on the right for each gene.