Figure 4. Non-conservative amino acid substitutions may contribute to constitutive activity in SmGAR.
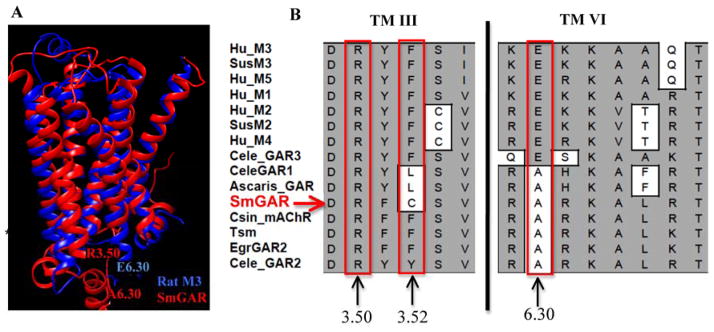
(A) A homology model of SmGAR (red) was superimposed onto the crystal structure of the rat M3 muscarinic receptor (4daj) (blue). The SmGAR model shows a typical topology of Family A GPCRs, including the canonical 7 transmembrane (TM) domains. Residues of the predicted “ionic lock” of Family A GPCRs are shown at the cytosolic ends of TM 3 and TM 6. Residue R3.50 corresponds to Arg2483.50 in SmGAR and Arg1653.50 in the rat M3 receptor. E6.30 refers to Glu4856.30 of the rat M3 receptor and A6.30 is the corresponding alanine in SmGAR (Ala8486.30). (B) Structural alignment of vertebrate and invertebrate muscarinic receptors showing a portion of TM 3 and TM6. The numbers describe the relative positions of amino acids of interest in each TM helix, according to the Ballasteros and Weinstein system [33], as described in the text. Mammalian muscarinic receptors are identified according to subtype, M1–M5; Invertebrate receptors are listed as GAR (G protein-coupled acetylcholine receptor) or mAChR (muscarinic acetylcholine receptor). Species abbreviations are as described in Fig. 1 and accession numbers are provided in Table S1.
