Abstract
The origin of protein import was a key step in the endosymbiotic acquisition of mitochondria. Though the main translocon of the mitochondrial outer membrane, TOM40, is ubiquitous among organelles of mitochondrial ancestry, the transit peptides, or N-terminal targeting sequences (NTSs), recognised by the TOM complex, are not. To better understand the nature of evolutionary conservation in mitochondrial protein import, we investigated the targeting behavior of Trichomonas vaginalis hydrogenosomal proteins in Saccharomyces cerevisiae and vice versa. Hydrogenosomes import yeast mitochondrial proteins even in the absence of their native NTSs, but do not import yeast cytosolic proteins. Conversely, yeast mitochondria import hydrogenosomal proteins with and without their short NTSs. Conservation of an NTS-independent mitochondrial import route from excavates to opisthokonts indicates its presence in the eukaryote common ancestor. Mitochondrial protein import is known to entail electrophoresis of positively charged NTSs across the electrochemical gradient of the inner mitochondrial membrane. Our present findings indicate that mitochondrial transit peptides, which readily arise from random sequences, were initially selected as a signal for charge-dependent protein targeting specifically to the mitochondrial matrix. Evolutionary loss of the electron transport chain in hydrogenosomes and mitosomes lifted the selective constraints that maintain positive charge in NTSs, allowing first the NTS charge, and subsequently the NTS itself, to be lost. This resulted in NTS-independent matrix targeting, which is conserved across the evolutionary divide separating trichomonads and yeast, and which we propose is the ancestral state of mitochondrial protein import.
Keywords: mitochondria, hydrogenosomes, mitosomes, protein import, TOM/TIM
Introduction
The origin of mitochondria marked the emergence of eukaryotes (Williams et al. 2013; McInerney et al. 2014), whose increased cellular complexity over prokaryotes is founded in the compartmentalization of chemiosmotic ATP (adenosine tri-phosphate) synthesis in the organelle (Martin and Koonin 2006; Lane and Martin 2010). All known eukaryotes possess, or possessed in their past, mitochondria or organelles derived thereof—hydrogenosomes and mitosomes (Van der Giezen et al. 2002). The family of mitochondrial organelles underwent different trajectories of specialization in different eukaryotic lineages. Aerobic mitochondria use O2 as the terminal electron acceptor, anaerobic mitochondria use other terminal acceptors such as fumarate. Hydrogenosomes generate ATP via H2-producing fermentations, while mitosomes consume ATP, rather than generating it (Muller et al. 2012). Hydrogenosomes are evolutionarily reduced in that they have lost the respiratory chain and the electrochemical gradient (Δψ) and instead generate ATP through substrate-level phosphorylation only (Lindmark and Muller 1973). Mitosomes are the most highly reduced forms of mitochondria, their only known functions involving Fe–S cluster assembly (Lill and Neupert 1996; Goldberg et al. 2008) and sulfur metabolism (Mi-ichi et al. 2009). Despite this specialization, mitochondrial protein import is conserved. Mitosomes of Encephalitozoon cuniculi might import only as few as 22 proteins (Katinka et al. 2001; Waller et al. 2009), yet like any other eukaryote studied so far, they depend on a mitochondrial translocon machinery consisting of components conserved in the canonical TIM and TOM complexes (Translocase of the Outer/Inner Mitochondrial membrane) of yeast and human mitochondria to do so (Doležal et al. 2006; Neupert and Herrmann 2007; Chacinska et al. 2009; Endo and Yamano 2009; Schleiff and Becker 2010).
Early in mitochondrial evolution, the invention of a protein import machinery allowed the organelle to relinquish genes to the nucleus (Timmis et al. 2004), but in order for the organelle to maintain its biochemical identity, and hence fulfill its bioenergetic functions, a mechanism that selectively discriminated between proteins germane to the organelle and pre-existing host proteins in the cytosol must have been in place. Today, this discrimination is provided by the TOM and TIM complexes, which comprise the core of the mitochondrial protein import machinery (Doležal et al. 2006; Chacinska et al. 2009; Schleiff and Becker 2010; Neupert 2015). In the oxygen-respiring mitochondria of yeast and humans, hundreds of matrix proteins enter the organelle via the TOM receptor platform that interacts with mitochondrial N-terminal targeting sequences (mNTSs) (Neupert and Herrmann 2007; Chacinska et al. 2009; Schleiff and Becker 2010). Anaerobic organelles of mitochondrial origin, hydrogenosomes and mitosomes, import fewer proteins than classical mitochondria but still make use of the same core components of the TOM and TIM machinery (Waller et al. 2009).
The main TOM component, Tom40, shuttles the unfolded preproteins into the inner membrane space, where they are received by the TIM23 complex that translocates proteins into the matrix in a process that in yeast requires both ATP and an electrochemical gradient (Δψ) across the inner membrane (Martin et al. 1991). Proteins targeted to the mitochondrial matrix harbor N-terminal targeting sequences (mNTSs) that can readily arise from random sequences (Baker and Schatz 1987) and that are present naturally in bacterial genomes (Lucattini et al. 2004). Although the translocases of the mitochondrial outer and inner membranes are ubiquitous among organelles of mitochondrial ancestry, positively charged NTSs that direct proteins to the organellar matrix are not (Regoes et al. 2005; Goldberg et al. 2008; Šmíd et al. 2008; Waller et al. 2009; Zimorski et al. 2013).
The two membranes that surround hydrogenosomes harbor many homologs of the TOM/TIM machinery. Proteins present include TOM40, TIM23, and proteins of the SAM and PAM complex, but they appear to lack many of the peripheral components of the mitochondrial targeting machinery as proteomic profiling has shown (Rada et al. 2011). Trichomonas hydrogenosomes lack a genome and therefore import all of the 200–500 proteins that exist in the organelle from the cytosol (Burstein et al. 2012). The Trichomonas genome encodes 226 proteins that harbor a short N-terminal motif with conserved features thought to represent the hydrogenosomal N-terminal targeting sequence or hNTS (Carlton et al. 2007; Burstein et al. 2012). This hNTS, while short, has been shown in some cases to be sufficient to target marker proteins to mitochondria of yeast (Häusler et al. 1997). Surprisingly, though, the deletion of the hNTS had only a marginal, if any, impact on the targeting efficiency of at least eight Trichomonas matrix proteins to hydrogenosomes (Mentel et al. 2008; Burstein et al. 2012; Zimorski et al. 2013), raising questions about the role and essentiality of the hNTS in hydrogenosomal protein import. To investigate the extent to which N-terminal independent targeting is conserved across the evolutionary divide that separates excavates and opisthokonts, we analyzed the targeting behavior of Trichomonas hydrogenosomal proteins in yeast and, reciprocally, the targeting of yeast mitochondrial proteins in Trichomonas with and without their NTSs.
Materials and Methods
Cultivation and Cloning
Trichomonas vaginalis T1 (J-H Tai, Institute of Biomedical Sciences, Taipei, Taiwan) was cultivated in TYM (Tryptone-Yeast extract-Maltose) medium at 37 °C as described previously (Gorrell et al. 1984). Saccharomyces cerevisiae INVSc1 was obtained from Invitrogen (Cat.No C810-00) and cultivated in YPD (Yeast extract-Peptone-Dextrose) (2% [w/v] glucose, 1% [w/v] yeast extract, and 2% [w/v] peptone) at 30 °C. Transfected yeast strains were cultivated in SC (Synthetic Complete) minimal medium (0.67% [w/v] yeast nitrogen base, 0.96% (w/v) yeast synthetic dropout medium without uracil) supplemented with 1% (w/v) raffinose. Open reading frames of the genes were cloned using genomic DNA of T. vaginalis T1 or genomic DNA of INVSc1 as template using gene-specific primers containing appropriate sites for the respective restriction enzymes as listed in supplementary table S2, Supplementary Material online. pTagVag2 was used for expression of genes in T. vaginalis which contains a C-terminal di-hemagglutinin (HA) tag and the promoter of the TvSCSα1 gene. For expression in S. cerevisiae, an inducible expression vector pYES2/eGFP (pYES2/CT with a C-terminal eGFP) was used whereby fusion constructs could be induced by the addition of 4% (w/v) galactose. Trichomonas vaginalis T1 cells were transfected as described before (Land et al. 2003) with 50 µg of plasmid and selected with 100 µg/ml G418. Transformation of S. cerevisiae cells was carried out using the protocol described in the manufacturer’s manual.
Cell Fractionation and Organelle Isolation
Isolations of hydrogenosomes were performed exactly as described before in Zimorski et al. (2013) except for an additional isopycnic centrifugation in 45% (v/v) Percoll density gradient with two intermediary washing steps to remove contaminating fractions. The isolation of mitochondria was carried out according to the protocol detailed in Gregg et al. (2009) with transfected cells grown in SC minimal medium. The total lysate fraction was collected immediately after the homogenization of cells. The supernatant of the pelleted mitochondria represents the cytoplasmic fraction.
Western Blotting, Immunofluorescence, and Imaging
Protein samples were separated through standard SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) procedures and blotted onto a nitrocellulose membrane. The membranes were blocked in 5% (w/v) dried milk powder in Tris-buffered saline with 0.1% (v/v) Tween20 (TBS-T) (tris-buffered-saline-tween20) for 30 min. The blots were incubated with primary antibodies at a concentration of 1:1,000 in blocking buffer overnight at 4 °C or 60 min at RT (room temperature) and then washed with TBS-T followed by incubation with secondary antibodies at a concentration of 1:5,000 in blocking buffer with 1% (w/v) dried milk powder for 60 min at RT and subsequent washes before imaging the blots directly in a Bio-Rad ChemiDoc™ XRS system.
For immunofluorescent labeling cells fixed in 1% (v/v) paraformaldehyde were deposited on cover slides coated with 0.01% polylysine and permeabilized for 15 min in 0.5% (v/v) Triton-X100. Permeabilized cells were blocked using a blocking buffer containing 1% (w/v) bovine serum albumin for 60 min followed by incubation in mouse anti-HA monoclonal antibody (Sigma) and rabbit anti-SCSα polyclonal serum in blocking buffer at a 1:1,000 dilution overnight at 4 °C. The cells were then washed three times in PBS before incubation with donkey antimouse Alexa 488 and donkey antirabbit Alexa 594 antibodies at a 1:5,000 dilution in blocking buffer for 60 min at RT. After final washes cells were mounted using Fluroshield™ containing DAPI (4′,6-diamidino-2-phenylindole) (Sigma) and observed using the Zeiss LSM710 confocal microscopy system.
Induction of eGFP (enhanced green fluroscent protein) fusion construct expressing yeast cells was carried out by growing log-phase yeast transformants in the presence of 4% (v/v) galactose for 4 h followed by incubation with 1 nM MitoTracker Red CMXRos (Invitrogen) and then mounted on silane coated slides in a solution of 1.2% (w/v) agarose to immobilize the cells and visualized in the Zeiss LSM710 confocal microscope. All images were analyzed using ImageJ software (Pérez and Pascau 2013).
Analysis of Hydrogenosomal Proteins by Liquid Chromatography-Electrospray Ionization MS/MS
Samples were digested and analyzed using liquid chromatography (LC)-electrospray ionization (ESI) mass spectrometry. Protein lysates (5 µg) were focused on a 4–12% polyacrylamide bis-tris gel (Life Technologies). After silver staining, protein bands were cut, destained (15 mM Na2S2O3, 50 mM K3[Fe(CN)6]), reduced (10 mM DTT (dithiothreitol), 50 mM (NH4)HCO3), alkylated (50 mM C2H4INO, 50 mM NH4HCO3), and proteins were digested overnight in 50 mM NH4HCO3, with 0.1 µg trypsin (Serva) or 0.1 µg GluC (Promega). Alternatively, digestion with 0.1 µg ArgC (Promega) was carried out in ArgC digestion buffer (50 mM Tris [pH 7.6], 2 mM ethylenediaminetetraacetic acid, 5 mM DTT, and 4.5 mM CaCl2). For LC-MS/MS (mass spectrometry) analyses, peptides were extracted from the gel with 1:1 (v/v) 0.1% TFA (trifluroacetic acid)/acetonitrile and after removal of acetonitrile 500 ng peptides were subjected to LC.
An Ultimate 3000 Rapid Separation liquid chromatography system (Dionex/Thermo Scientific) was used for peptide separation. After injection, peptides were preconcentrated on an Acclaim PepMap100 trap column (3 µm C18 particle size, 100 Å pore size, 75 µm inner diameter, 2 cm length; Dionex/Thermo Scientific) at a flow rate of 6 µl/min using 0.1% (v/v) TFA as mobile phase. After 10 min, peptides were separated on an analytical column (Acclaim PepMapRSLC, 2 µm C18 particle size, 100 Å pore size, 75 µm inner diameter, 25 cm length; Dionex/Thermo Scientific) at 60 °C using a 2-h gradient from 4% to 40% solvent B (solvent A: 0.1% (v/v) formic acid in water’ solvent B: 0.1% (v/v) formic acid, 84% (v/v) acetonitrile in water) at a flow rate of 300 nl/min.
Mass spectrometry was carried out on an Orbitrap Elite high resolution instrument (Thermo Scientific) operated in positive mode and equipped with a nano ESI source. Capillary temperature was set to 275 °C and source voltage to 1.4 kV. Survey scans were carried out in the orbitrap analyzer over a mass range from 350 to 1,700 m/z at a resolution of 60,000 (at 400 m/z). The target value for the automatic gain control was 1,000,000 and the maximum fill time was 200 ms. The 20 most intense 2+ and 3+ charged peptide ions (minimal signal intensity 500) were isolated, transferred to the linear ion trap (LTQ [linear trap quadrupole]) part of the instrument, and fragmented using collision induced dissociation. Peptide fragments were analyzed with a maximal fill time of 300 ms and automatic gain control target value of 10,000. The available mass range was 200–2,000 m/z at a resolution of 5,400 (at 400 m/z). Already fragmented ions were excluded from fragmentation for 45 s.
Analysis of Mass Spectrometric Data
Raw files were processed with MaxQuant (version 1.4.1.2, Max Planck Institute for Biochemistry, Munich, Germany) for protein and peptide identification and quantification with default parameters if not otherwise stated. Searches were carried out using T. vaginalis protein sequences from TrichDB (release 1.3 from 26.5.2011 including 59,672 protein entries; Aurrecoechea et al. 2008) applying the following parameters: Mass tolerance precursor (Orbitrap): 20 ppm first search, 4.5 ppm second search; mass tolerance fragment spectra (linear ion trap): 0.5 Da (linear ion trap); fixed modification: Carbamidomethyl (C), nicotin (K); variable modifications: Mthionine oxidation. Searches with protease-specific cleavage (depending on the used enzyme GluC, ArgC, maximum of two missed cleavage sites) were used with specific cleavage and in an alternative setting with N-terminal semispecific cleavage.
For peptide and protein acceptance, the false discovery rate (FDR) was set to 1%, and only proteins with at least two identified peptides were used for protein assembly. Quantification was carried out using the label-free quantification algorithm implemented in MaxQuant using a minimal ratio count of 2 and the “match between runs” option enabled. Alternatively, raw files were further processed for protein and peptide identification using Proteome Discoverer (version 1.4.1.14, Thermo Scientific) connected to a Mascot server (version 2.4.1, Matrix Sciences, London, UK) with default parameters for spectrum selection. Searches were carried out using 59,672 protein entries and protein sequences from TrichDB (release 1.3 from 26.5.2011) applying the following parameters: Mass tolerance precursor (analyzed in the Orbitrap part of the instrument) 10 ppm, mass tolerance fragment spectra (analyzed in the linear ion trap) 0.4 Da, enzyme-specific cleavage with a maximum of one missed cleavage site and N- and C-terminal semispecific cleavage specificity, carbamidomethyl at cysteines and nicotine at lysines as fixed modification and methionine oxidation. For peptide and protein acceptance, the “Percolator” function with a Target FDR set to 1% and validation based on q-value was used. Only peptides with high confidence (FDR < 1%) were used for protein assembly. Protein grouping was enabled. Net charge of peptides was analyzed using the EMBOSS package pepstats (Rice et al. 2000).
Results
The Majority of Hydrogenosomal Proteins Do Not Harbor an N-Terminal Targeting Sequence
A previous proteomic investigation of isolated hydrogenosomes from T. vaginalis identified 536 proteins, including 99 proteins for which only one peptide was identified (Schneider et al. 2011). Hydrogenosomes thus harbor on the order of 4–500 proteins, which is about half as many proteins as are predicted to localize to yeast mitochondria (Meisinger et al. 2008), but about twice the number of Trichomonas proteins (226) predicted by Burstein et al. (2012) to contain an hydrogenosomal NTS. A subsequent proteomic study of only the membrane associated proteins (Rada et al. 2011) revealed another 102 proteins that were not identified by Schneider et al. (2011). This prompted us to reinvestigate the Trichomonas hydrogenosomal proteome. Using biological triplicates of highly purified isolated hydrogenosomes and proteolytic digestion of the isolated proteins by two independent proteases, ArgC and GluC, for analysis by mass spectrometry (supplementary table S1, Supplementary Material online), 359 proteins were common to the three separate LC-MS/MS runs and detected with at least two peptides per protein. Of these 359 proteins, only 39 proteins have an hNTS based on previous predictions (Burstein et al. 2012; fig. 1A). The rest of the 320 proteins lacked a predictable hNTS altogether, including proteins that were previously not identified (supplementary table S2, Supplementary Material online).
Fig. 1.—
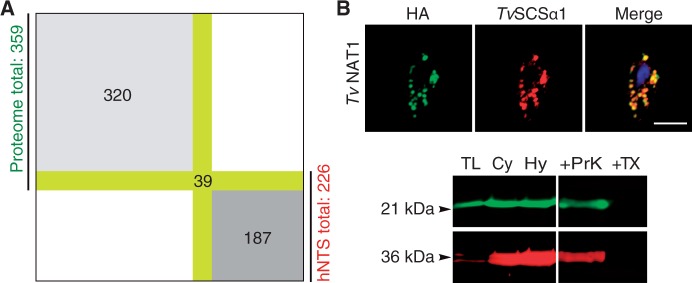
The majority of hydrogenosomal proteins lack an hNTS. (A) Of the 226 proteins predicted to have an hNTS, only 39 were found in the hydrogenosomal proteome of 359 proteins identified in total. (B) Immunofluorescent colocalization of the HA-tagged TVAG_270750 that harbors no hNTS and the hydrogenosomal marker enzyme TvSCSα (succinyl coenzyme A synthetase). Below a multiplex western blot of the protease protection assay on isolated hydrogenosomes with green representing anti-HA and red anti-TvSCSα. TL, total lysate; Cy, cytosol; Hy, hydrogenosomes; TX, 0.1% Triton X-100; ProtK, Triton X-100 + 100 µg/µl proteinase K. Numbers to the left indicate the approximate molecular weights of the constructs in kilo Daltons (kDa).
One of those proteins lacking an hNTS was TVAG_270750 (TvNAT1), which shares ∼50% amino acid identity with bacterial acetyltransferases from the GNAT family. To confirm that TvNAT1 is a hydrogenosomal protein in vivo, we expressed TvNAT1 as an HA-tagged construct. It colocalized with TvSCSα1 (fig. 1B), a marker enzyme of the hydrogenosomal matrix (Zimorski et al. 2013). Matrix localization was further supported by a protease protection assay (PPA) on isolated hydrogenosomes (fig. 1B). TvNAT1 is thus yet one more in a growing list of proteins that localize to the Trichomonas hydrogenosomal matrix in the absence of an NTS (Mentel et al. 2008; Burstein et al. 2012; Zimorski et al. 2013). This prompted us to undertake a broader and more systematic investigation of NTS-independent targeting to hydrogenosomes.
Reciprocal Targeting of Mitochondrial and Hydrogenosomal Proteins with and without NTSs
We investigated the targeting behavior of T. vaginalis hydrogenosomal proteins in S. cerevisiae with and without their hNTSs. Four different hydrogenosomal matrix proteins of T. vaginalis (TvSCSα1, TvFdx, TvME, TvISCA1), whose hNTS had been previously shown to be nonessential in Trichomonas (Zimorski et al. 2013), were fused to the N-terminus of eGFP and localized in yeast. All four proteins of the parasite carrying their hNTS were targeted to yeast mitochondria (fig. 2A–D). These four proteins were also targeted to the mitochondria of S. cerevisiae when the proteins were expressed without their NTS (fig. 2A–D). As further support, mitochondria of the transformed strains were isolated and the subcellular fractions investigated in multiplex western blots using an anti-eGFP antibody and an antibody against CoxIV, a protein of the inner mitochondrial membrane (fig. 2A–E). The western blots confirmed the localization observed by immunofluorescent microscopy, that is, the fusion proteins were exclusively detected in the fractions containing either total protein or the proteins of the isolated yeast mitochondria, and that no matter of whether the hNTS was present or not.
Fig. 2.—
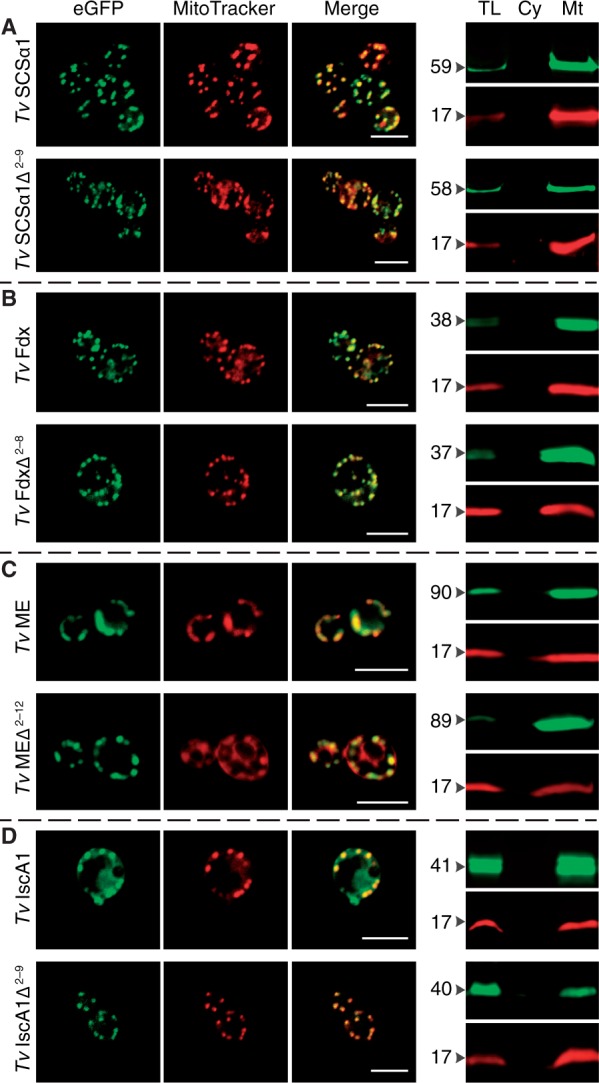
Hydrogenosomal proteins are targeted to yeast mitochondria with and without N-terminal leaders. (A–D) All four hydrogenosomal proteins of Trichomonas vaginalis analyzed (TvSCSα1, succinyl coenzyme A synthetase; TvFdx, ferredoxin; TvME, malic enzyme; TvISCA1; iron–sulfur assembly protein 1) are targeted to the mitochondria of yeast, even in the absence of their NTSs. Δ indicates the positions of the N-terminal amino acids (i.e., the hNTS) that were deleted. (E) Trichomonas actin was used as a control next to the transfection of the empty vector that expresses GFP alone (supplementary fig. S2, Supplementary Material online). Scale indicates 5 µm. On the right, the multiplex western blots of isolated mitochondria probed with anti-GFP antibody (green) and the mitochondrial marker protein anti-COXIV (red). TL, total lysate; Cy, cytosol; Mt, mitochondria. Numbers to the left indicate the approximate molecular weights of the constructs in kilo Daltons (kDa).
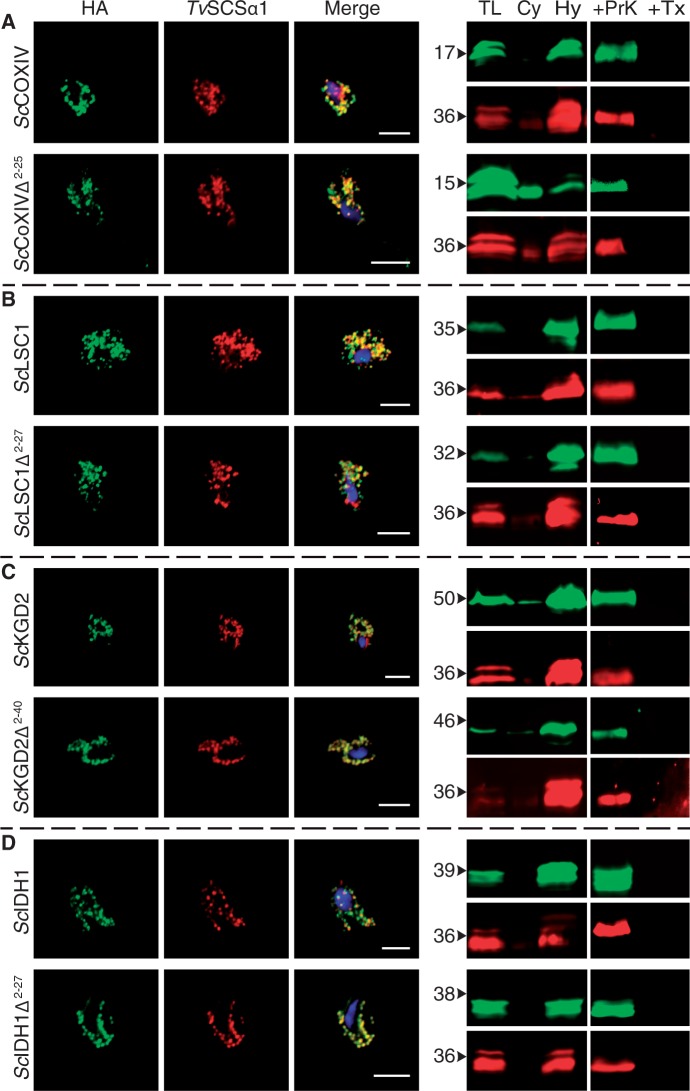
We also tested the reciprocal case. To determine if the converse was true for yeast mitochondrial proteins, we expressed four canonical and abundant yeast mitochondrial proteins (ScLSC1, ScCOXIV, ScIDH1, and ScKGD2) in the parasite T. vaginalis. ScLSC1 is the yeast homolog of TvSCSα1, ScCOXIV is part of the mitochondrial respiratory chain, and ScIDH1 and ScKGD2 are metabolic enzymes of the mitochondrial matrix for which Trichomonas encodes no homologs. An earlier study of the N-proteome of the mitochondria of yeast identified all four proteins to be present in the mitochondria (Vögtle et al. 2009). Moreover, these proteins were present in a processed form having their mNTS cleaved by a peptidase, which strongly suggests that their mNTS is necessary for mitochondrial import. All four yeast proteins localized to T. vaginalis hydrogenosomes independent of the presence or absence of their mNTS (fig. 3A–D). In addition, multiplex western blots of purified hydrogenosomes and subsequent PPAs were also performed and demonstrated the proteins to be present in the organellar fractions (fig. 3A–D).
Fig. 3.—
Yeast mitochondrial proteins are targeted to Trichomonas vaginalis hydrogenosomes with and without N-terminal leaders. (A–D) Expression of four mitochondrial proteins of Saccharomyces cerevisiae (ScLSC1, succinate coenzyme A ligase; ScCOXIV, cytochrome C oxidase; ScIDH1, isocitrate dehydrogenase; ScKGD2, alpha-ketoglutarate dehydrogenase) tagged with HA demonstrate that they are all targeted to hydrogenosomes in the presence and absence of their mNTS. Δ indicates the positions of the N-terminal amino acids (i.e., the mNTS) that were deleted. In the merge the DNA is stained through DAPI. Scale bar indicates 5 µm. On the right, multiplex western blots on isolated hydrogenosomes (TL, total lysate; Cy, Cytosol, Hy; hydrogenosomes) are shown along with a protease protection assay (TX, 0.1% TritonX-100; ProtK, Triton X-100 + 100 µg/µl proteinase K). Green bands represent the HA-tag, red bands TvSCSα1. Numbers to the left indicate approximate molecular weights of the constructs in kilo Daltons (kDa).
This targeting and localization is restricted to proteins of organellar origin in both organisms. As controls for yeast, we localized eGFP alone and additionally fused the eGFP to the C-terminus of an actin gene of T. vaginalis (TvActin). Both constructs remained in the cytosol and did not colocalize with MitoTracker® Red (fig. 4A). Three cytosolic yeast proteins (the glycolytic enzymes ScGAPDH, ScActin, and ScRab5) were expressed in the parasite as a control using the same expression vector that is pTagVag2. The fusion proteins did not associate with the hydrogenosomes of Trichomonas (fig. 4B). This further demonstrates that the recognition of import substrate at the hydrogenosomal and mitochondrial outer membrane is specific, even in the absence of an NTS. The data indicate that some internal targeting information must exist in these yeast mitochondrial and Trichomonas hydrogenosomal proteins recognized by both the yeast and Trichomonas organelle protein import machinery that can discriminate between cytosolic proteins and proteins of the organelle. The nature of that targeting information remains obscure.
Fig. 4.—
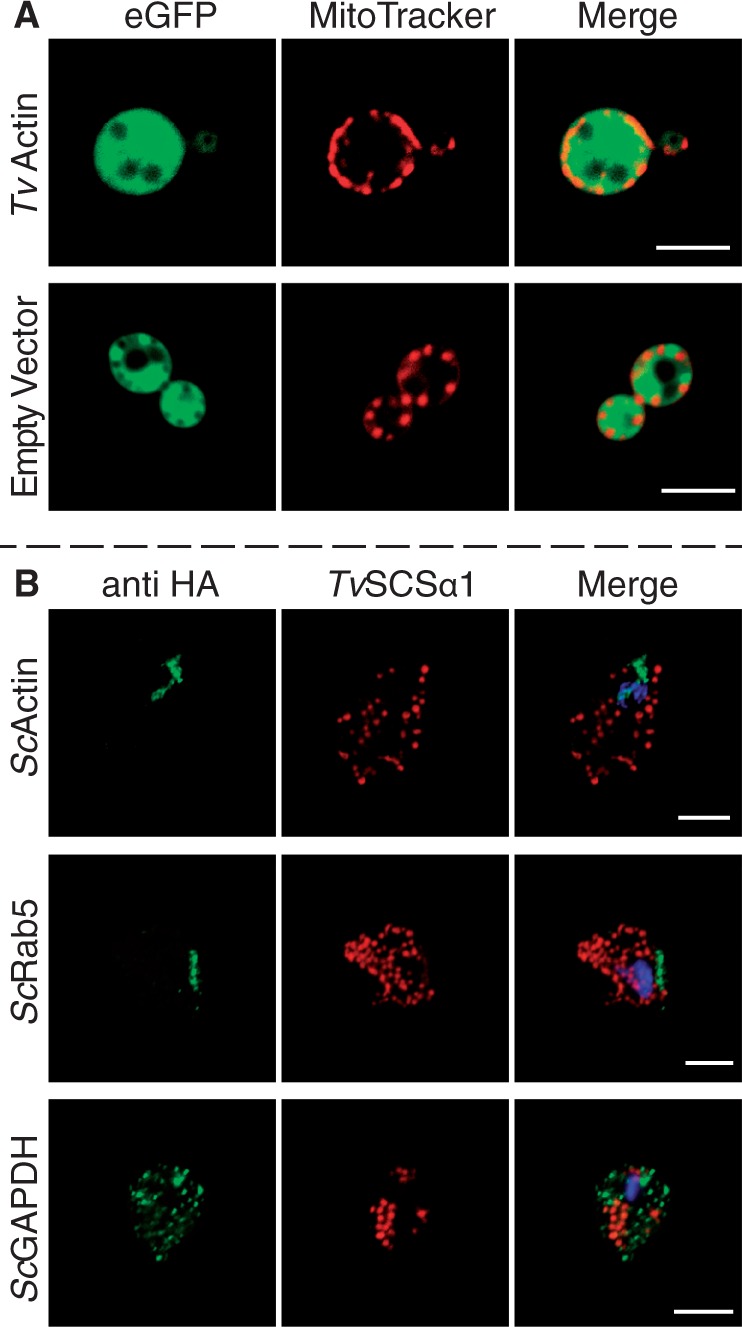
Cytosolic proteins do not colocalize with the organelles. (A) Expression of TvActin (Trichomonas actin) and the empty vector expressing the eGFP tag alone demonstrates that they do not localize to the mitochondria stained with MitoTracker® Red. (B) Three cytosolic proteins from yeast (ScGAPDH, glyceraldehyde-3-phosphate dehydrogenase; ScActin, actin; ScRab5, rab family GTPase) were expressed using the same expression vector and yet do not colocalize with the hydrogenosomal marker TvSCSα1.
Discussion
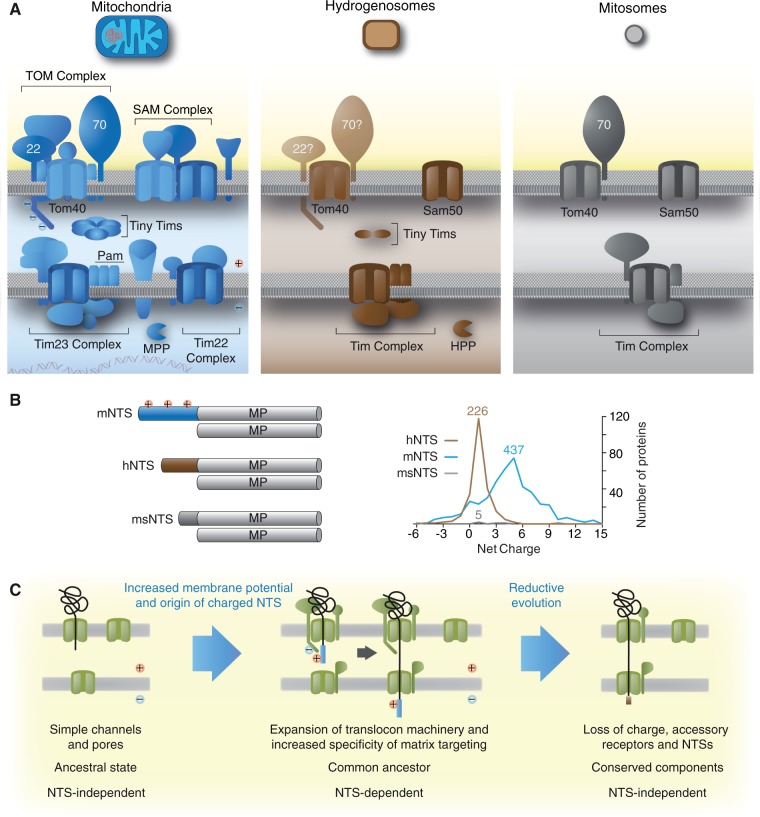
Organelles of mitochondrial origin share a common ancestry (Muller et al. 2012; Makiuchi and Nozaki 2014). In some eukaryotes, such as Trichomonas and Giardia, the organelles have undergone reduction to become hydrogenosomes and mitosomes, respectively. This process is accompanied by the loss of oxidative phosphorylation coupled with a loss of the electrochemical gradient, Δψ, across the inner membrane, loss of the organellar genome and translation machinery, and a reduction in the number of proteins that are targeted to the organelles (Muller 1993; Goldberg et al. 2008; Jedelský et al. 2010; Schneider et al. 2011). Accompanying that biochemical and functional reduction, the protein import machinery has also undergone reduction from a very complex receptor platform in mitochondria to a more minimalistic import machinery in mitosomes (Doležal et al. 2006; Waller et al. 2009; Schleiff and Becker 2010). Targeting of matrix proteins to mitochondria is initiated through NTSs that are recognized by receptors, which are associated with the outer membrane of the organelle (Schleiff and Becker 2010). Although this process is conserved across all eukaryotes, the nature of the translocon machinery operating in the very earliest eukaryotes is still obscure.
Current views have it that the core translocons of TOM and TIM trace back to prokaryotic membrane proteins (Hewitt et al. 2011) and were hence present in the ancestor of mitochondria. A general analysis of eukaryotic porins revealed that Tom40 shares a significant structural homology with the beta-barrel structure of bacterial porins (Zeth and Thein 2010) and that the main translocation pores of the TIM complex, Tim23 and Tim17, evolved from common bacterial transporters (Rassow et al. 1999). In contrast to Tom40, which is highly conserved, Tom20 is far more variable than the core translocases and might have even evolved several times independently (Perry et al. 2006). That in turn suggests that the receptors for the NTS, although ubiquitous among eukaryotes, evolved after the origin of translocation pores of the two import complexes TOM and TIM, which were thus ancestral.
Essential components of TOM and TIM are conserved in hydrogenosomes and mitosomes (Regoes et al. 2005; Doležal et al. 2006; Rada et al. 2011). Mitosomes are even more reduced than hydrogenosomes (Waller et al. 2009; Heinz and Lithgow 2013), the organelles of E. cuniculi import only a few dozen proteins (Katinka et al. 2001; Waller et al. 2009), but also employ conserved TOM and TIM components (Waller et al. 2009). Similar to the situation with Trichomonas hydrogenosomes, mitosomal NTSs, when present, are short, with the majority of proteins targeted to microsporidian mitosomes lacking N-terminal extensions altogether (Katinka et al. 2001; Waller et al. 2009). Earlier findings that Trichomonas proteins localize to hydrogenosomes independent of their short hNTSs (Mentel et al. 2008; Burstein et al. 2012; Zimorski et al. 2013), along with similar observations for Giardia (Regoes et al. 2005) and more recently Trypanosoma (Hamilton et al. 2014), indicate that internal motifs of yet unknown nature can interact with the TOM translocon and mediate subsequent translocation of the organellar proteins without the need for an NTS. For those hydrogenosomal and mitosomal proteins, which have retained an NTS, the net positive charge—a conserved hallmark of mitochondrial NTSs (von Heijne 1986)—is lost (fig. 5B).
Fig. 5.—
The complexity of the mitochondrial targeting machinery reduces together with the evolutionarily reduction of the organelle’s biochemistry. (A) An illustration of the organellar import machineries from mitochondria, hydrogenosomes, and mitosomes. It is estimated that classical mitochondria import between 1,000 and 1,500 proteins, while hydrogenosomes of Trichomonas import only 200–500 proteins. (B) Mitochondrial matrix proteins carry long NTSs that are usually positively charged, but a few lack a recognizable mNTS. If present, the hydrogenosomal NTS is shorter and no longer charged. Mitosomal proteins of Encephalitozoon cuniculi completely lost the N-terminal targeting information and solely rely on internal motifs. Mitosomes of E. cuniculi are streamlined to such a degree, where they might require to import as little as 22 proteins from the cytosol; proteins that are largely responsible for the last known remaining biochemical pathway, which is iron–sulfur cluster (Fe–S) biosynthesis. (C) Early during mitochondrial evolution, transmembrane protein import was NTS independent. Import complexity (amount of proteins involved and the required targeting information) evolved downstream. During reductive evolution of the organelle, for instance in anaerobic parasites, import complexity decreased back, toward NTS-independent recognition of organellar proteins, translated in the cytosol.
That the hNTS is not required for hydrogenosomal targeting is supported by our proteome analysis (supplementary table S1, Supplementary Material online). In 6 separate LC-MS/MS runs that were based on biological triplicates, 359 proteins were identified with a minimum of 2 peptides per protein. One hundred eighty-seven of the proteins that harbor a predicted NTS—and thus were good candidates to constitute the core hydrogenosomal proteome—were absent from our analysis. They might be of low abundance, not expressed at all, or further evidence that the correct targeting to hydrogenosomes does not hinge upon the presence of that very hNTS. Three hundred twenty proteins (∼90% of the hydrogenosomal proteins identified) lacked an hNTS (fig. 1). The lack of NTS is not without precedent in mitochondria. A global N-proteome of yeast mitochondria identified 400 proteins with processed N-termini of at least 10 a.a. (amino acids) out of 585 proteins identified in total (Vögtle et al. 2009). Although many yeast mitochondrial proteins are directed to the inner and outer membranes as well as to the intermembrane space (IMS) without an NTS, targeting to the yeast mitochondrial matrix appears to be NTS dependent in cases reported so far (Neupert and Herrmann 2007; Chacinska et al. 2009; Schleiff and Becker 2010; Neupert 2015).
Our present data (summarized in table 1) provide more evidence that in Trichomonas hydrogenosomal targeting works in the absence of an NTS, albeit some might still require it (Mentel et al. 2008; Zimorski et al. 2013). In any case, this mode of NTS-independent targeting is—at least for the proteins tested—conserved in yeast, because hydrogenosomal matrix proteins lacking an NTS are directed to the yeast mitochondrion (fig. 2). Moreover, the converse is true of yeast matrix proteins in Trichomonas hydrogenosomes (fig. 3). Vector-caused localization artifacts can be ruled out. Our controls (empty vector and fusion proteins involving nonorganellar proteins of Trichomonas and yeast) never colocalized with hydrogenosomal markers (fig. 4). In addition, the vector used for the transfection of Trichomonas was previously used to analyze surface proteins (Noël et al. 2010), nuclear proteins (Zubácová et al. 2012), and cytoskeletal proteins (Kusdian et al. 2013). None of these fusion proteins associated with the hydrogenosomes. The same is true for yeast and the pYES2/CT plasmid (Donahue et al. 2001; Todisco et al. 2014). In summary, this indicates that proteins of mitochondrial ancestry have yet unspecified properties that mediate interactions with the Tom40 translocon, which was present in the earliest eukaryotes. The nature or identity of these properties remains so far unidentified.
Table 1.
Summary of the Localization Studies
| Gene | NTS (a.a.) | Localization in Trichomonas vaginalis |
Localization in S. cerevisiae |
|||
|---|---|---|---|---|---|---|
| Full | Δm/hNTS | Full | Δm/hNTS | |||
| S. cerevisiae | ScKGD2 | 40 | Hy | Hy | Mt | Uk |
| ScIDH1 | 11 | Hy | Hy | Mt | Uk | |
| ScCOXIV | 25 | Hy | Hy | Mt | Cy | |
| ScLSC1 | 27 | Hy | Hy | Mt | Uk | |
| ScACT1 | — | Ct | — | Ct | — | |
| ScGAPDH | — | Cy | — | Cy | — | |
| ScRab5 | — | Cy/En | — | En | — | |
| T. vaginalis | TvSCSα1 | 9 | Hy | Hy | Mt | Mt |
| TvME | 12 | Hy | Hy | Mt | Mt | |
| TvISCA1 | 9 | Hy | Hy | Mt | Mt | |
| TvFdx | 8 | Hy | Hy | Mt | Mt | |
| TvActin | — | Ct | — | Cy | — | |
| eGFP | — | — | — | Cy | — | |
Note.—The length of the NTS was determined using TargetP (Emanuelsson et al. 2000). Hy, hydrogenosomes; Mt, mitochondria; Cy, cytosol; Ct, cytoskeleton; En, endosomes; Uk, unknown.
This raises a curious question: If Tom40 can recognize its own substrates, why did NTS-dependent targeting evolve in the first place, and more intriguingly, why is it preferentially lost in hydrogenosomes and mitosomes? One possible rationale for the origin of NTS-dependent targeting is specificity. The presence of a dedicated receptor-ligand (TOM-NTS) pair for recognition and import would allow increased specificity of TOM interactions and thus channel substrates to the TIM complex. Although the origin of a sophisticated receptor platform including Tom20, Tom22, and Tom70 (fig. 4A) might have been selected for NTS recognition and specificity, import of proteins lacking an NTS is also specific (Regoes et al. 2005; Goldberg et al. 2008; Mentel et al. 2008; Šmíd et al. 2008; Waller et al. 2009; Burstein et al. 2012; Zimorski et al. 2013; Hamilton et al. 2014). Hence, receptor interactions at the TOM complex alone cannot explain the presence of an NTS. We suggest that conservation of NTS-independent targeting of yeast and trichomonad proteins to the organelle constitute conserved, not convergent properties, and that they reflect the ancestral state of mitochondrial protein recognition and import from the cytosol.
It is well established that mitochondrial membrane potential electrophoretically directs the NTS to the TIM channel via the negatively charged tail of Tom22 (Pfanner and Neupert 1986; Martin et al. 1991; Esaki et al. 2004). In accordance with the “increasing affinity” model (Schleiff and Becker 2010), Tom22 binds the positively charged NTS within the IMS and recruits the TIM complex and TOM and TIM form a continuous pore across both membranes (Schleiff and Becker 2010). Noncleavable internal motifs target proteins to the mitochondrial IMS and the membranes (Chacinska et al. 2009; Schleiff and Becker 2010). In case of IMS proteins that have a charged mNTS, like cytochrome b2, the mNTS needs to traverse the matrix first and manipulation of the charged region of the mNTS decreases import efficiency (Geissler et al. 2000). Indeed in some cases, cytochrome b2 destined to the IMS lacks an NTS altogether (Hewitt et al. 2012).
We propose that in the eukaryotic common ancestor, a positively charged NTS was initially selected at the termini of matrix proteins for their electrophoretic import via the membrane potential across the inner membrane (fig. 5C), providing specific targeting to their designated compartment—the matrix. The evolution of the positive charge on the NTS also allowed discrimination between mitochondrial matrix and nonmatrix destinations. The ease with which a functional NTS can be obtained through random DNA sequences (Baker and Schatz 1987) indicates that the evolutionary origin of transit peptides was facile, requiring virtually no innovation at all (Lucattini et al. 2004), merely selection for accrual of positive charges on the N-termini of matrix-specific proteins and for subsequent proteolytic processing via the conserved mitochondrial processing peptidase (Šmíd et al. 2008).
The simpler nature of protein import in hydrogenosomal and mitosomal evolution has often, and rightly, been attributed to the general process of reductive evolution (Van der Giezen et al. 2002; Doležal et al. 2006; Šmíd et al. 2008). Our proposal that positive charge on the NTS arose as a matrix-specific targeting signal suggests what, exactly, was lost first (the charge), while uncovering the existence and conservation—though not the nature—of NTS-independent import signals in Trichomonas and yeast. Loss of the electron transport chain in the inner membrane in hydrogenosomes (and mitosomes) led to loss of Δψ, rendering positive charge on the NTS superfluous, hence readily lost through mutation. This accounts for the conspicuous lack of charge in hydrogenosomal and mitosomal NTSs. In the absence of charge, the NTS itself could however only become expendable in the event that either 1) a novel NTS-independent import pathway arose in the inner membrane in a lineage specific manner or 2) a conserved import pathway pre-existed that accommodated NTS-independent import. Conservation of NTS-independent targeting in Trichomonas and yeast indicate that the latter was the case. Our results bring into question the prevalence, evolutionary conservation, and antiquity of internal or cryptic signals in proteins targeted to mitochondrial organelles.
Supplementary Materials
Supplementary tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Financial support through the Deutsche Forschungsgemeinschaft to S.B.G. (GO1825/4-1), an ERC grant to W.F.M. (666053) and the Czech Science Foundation (13-09208J) to J.T. is gratefully acknowledged. We thank Kai Stühler and Gereon Poschmann of the BMFZ HHU-Düsseldorf for their excellent support and help analyzing the proteome data.
Literature Cited
- Aurrecoechea C, et al. 2008. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 37:D526–D530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A, Schatz G. 1987. Sequences from a prokaryotic genome or the mouse dihydrofolate reductase gene can restore the import of a truncated precursor protein into yeast mitochondria. Proc Natl Acad Sci U S A. 84:3117–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, et al. 2012. A machine learning approach to identify hydrogenosomal proteins in Trichomonas vaginalis. Eukaryot Cell. 11:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležal P, Likic V, Tachezy J, Lithgow T. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313:314–318. [DOI] [PubMed] [Google Scholar]
- Donahue SL. 2001. Mitochondrial DNA ligase function in Saccharomyces cerevisiae. Nucleic Acids Res. 29:1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K. 2009. Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta. 1803:706–714. [DOI] [PubMed] [Google Scholar]
- Esaki M, et al. 2004. Mitochondrial protein import. J Biol Chem. 279:45701–45707. [DOI] [PubMed] [Google Scholar]
- Geissler A, et al. 2000. Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b(2) modulates the deltapsi-dependence of translocation of the matrix-targeting sequence. Mol Biol Cell. 11:3977–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AV, et al. 2008. Localization and functionality of microsporidian iron–sulphur cluster assembly proteins. Nature 452:624–628. [DOI] [PubMed] [Google Scholar]
- Gorrell TE, Yarlett N, Müller M. 1984. Isolation and characterization of Trichomonas vaginalis ferredoxin. Carlsberg Res Commun. 49:259–268. [Google Scholar]
- Gregg C, Kyryakov P, Titorenko VI. 2009. Purification of mitochondria from yeast cells. J Vis Exp. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton V, Singha UK, Smith JT, Weems E, Chaudhuri M. 2014. Trypanosome alternative oxidase possesses both an N-terminal and internal mitochondrial targeting signal. Eukaryot Cell. 13:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler T, Stierhof YD, Blattner J, Clayton C. 1997. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur J Cell Biol. 73:240–251. [PubMed] [Google Scholar]
- Heinz E, Lithgow T. 2013. Back to basics: a revealing secondary reduction of the mitochondrial protein import pathway in diverse intracellular parasites. Biochim Biophys Acta. 1833:295–303. [DOI] [PubMed] [Google Scholar]
- Hewitt V, Alcock F, Lithgow T. 2011. Minor modifications and major adaptations: the evolution of molecular machines driving mitochondrial protein import. Biochim Biophys Acta. 1808:947–954. [DOI] [PubMed] [Google Scholar]
- Hewitt VL, et al. 2012. A model system for mitochondrial biogenesis reveals evolutionary rewiring of protein import and membrane assembly pathways. Proc Natl Acad Sci U S A. 109:E3358–E3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedelský PL, et al. 2010. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One 6:e17285–e17285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450–453. [DOI] [PubMed] [Google Scholar]
- Kusdian G, Woehle C, Martin WF, Gould SB. 2013. The actin-based machinery of Trichomonas vaginalis mediates flagellate-amoeboid transition and migration across host tissue. Cell Microbiol. 15:1707–1721. [DOI] [PubMed] [Google Scholar]
- Land KM, et al. 2003. Targeted gene replacement of a ferredoxin gene in Trichomonas vaginalis does not lead to metronidazole resistance. Mol Microbiol. 51:115–122. [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467:929–934. [DOI] [PubMed] [Google Scholar]
- Lill R, Neupert W. 1996. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 6:56–61. [DOI] [PubMed] [Google Scholar]
- Lindmark DG, Muller M. 1973. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem. 248:7724–7728. [PubMed] [Google Scholar]
- Lucattini R, Likic VA, Lithgow T. 2004. Bacterial proteins predisposed for targeting to mitochondria. Mol Biol Evol. 21:652–658. [DOI] [PubMed] [Google Scholar]
- Makiuchi T, Nozaki T. 2014. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie. 100:3–17. [DOI] [PubMed] [Google Scholar]
- Martin J, Mahlke K, Pfanner N. 1991. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J Biol Chem. 266:18051–18057. [PubMed] [Google Scholar]
- Martin W, Koonin EV. 2006. Introns and the origin of nucleus—cytosol compartmentalization. Nature 440:41–45. [DOI] [PubMed] [Google Scholar]
- McInerney JO, O'Connell MJ, Pisani D. 2014. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat Rev Microbiol. 12:449–455. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Sickmann A, Pfanner N. 2008. The mitochondrial proteome: from inventory to function. Cell 134:22–24. [DOI] [PubMed] [Google Scholar]
- Mentel M, Zimorski V, Haferkamp P, Martin W, Henze K. 2008. Protein import into hydrogenosomes of Trichomonas vaginalis involves both N-terminal and internal targeting signals: a case study of thioredoxin reductases. Eukaryot Cell. 7:1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-ichi F, Abu Yousuf M, Nakada-Tsukui K, Nozaki T. 2009. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A. 106:21731–21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. 1993. The hydrogenosome. J Gen Microbiol. 139:2879–2889. [DOI] [PubMed] [Google Scholar]
- Muller M, et al. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 76:444–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. 2015. A perspective on transport of proteins into mitochondria: a myriad of open questions. J Mol Biol. 427:1135–1158. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. 2007. Translocation of proteins into mitochondria. Annu Rev Biochem. 76:723–749. [DOI] [PubMed] [Google Scholar]
- Noël CJ, et al. 2010. Trichomonas vaginalis vast BspA-like gene family: evidence for functional diversity from structural organisation and transcriptomics. BMC Genomics 11:99–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez JMM, Pascau J. 2013. Image processing with ImageJ. Packt Publishing Ltd. [Google Scholar]
- Perry AJ, Hulett JM, Likic VA, Lithgow T, Gooley PR. 2006. Convergent evolution of receptors for protein import into mitochondria. Curr Biol. 16:221–229. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Neupert W. 1986. Transport of F1-ATPase subunit beta into mitochondria depends on both a membrane potential and nucleoside triphosphates. FEBS Lett. 209:152–156. [DOI] [PubMed] [Google Scholar]
- Rada P, et al. 2011. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PLoS One 6:e24428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Dekker PJT, van Wilpe S, Meijer M, Soll J. 1999. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 286:105–120. [DOI] [PubMed] [Google Scholar]
- Regoes A, et al. 2005. Protein import, replication, and inheritance of a vestigial mitochondrion. J Biol Chem. 280:30557–30563. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Schleiff EE, Becker TT. 2010. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol. 12:48–59. [DOI] [PubMed] [Google Scholar]
- Schneider RE, et al. 2011. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int J Parasitol. 41:1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmíd O, et al. 2008. Reductive evolution of the mitochondrial processing peptidases of the unicellular parasites Trichomonas vaginalis and Giardia intestinalis. PLoS Pathog. 4:e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5:123–135. [DOI] [PubMed] [Google Scholar]
- Todisco S, et al. 2014. The Saccharomyces cerevisiae gene YPR011c encodes a mitochondrial transporter of adenosine 5′-phosphosulfate and 3′-phospho-adenosine 5′-phosphosulfate. Biochim Biophys Acta. 1837:326–334. [DOI] [PubMed] [Google Scholar]
- Van der Giezen M, et al. 2002. Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J. 21:572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, et al. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139:428–439. [DOI] [PubMed] [Google Scholar]
- von Heijne G. 1986. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5:1335–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, et al. 2009. Evidence of a reduced and modified mitochondrial protein import apparatus in microsporidian mitosomes. Eukaryot Cell. 8:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Cox CJ, Embley TM. 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504:231–236. [DOI] [PubMed] [Google Scholar]
- Zeth K, Thein M. 2010. Porins in prokaryotes and eukaryotes: common themes and variations. Biochem J. 431:13–22. [DOI] [PubMed] [Google Scholar]
- Zimorski V, et al. 2013. The N-terminal sequences of four major hydrogenosomal proteins are not essential for import into hydrogenosomes of Trichomonas vaginalis. J Eukaryot Microbiol. 60:89–97. [DOI] [PubMed] [Google Scholar]
- Zubácová Z, Hostomská J, Tachezy J. 2012. Histone H3 variants in Trichomonas vaginalis. Eukaryot Cell. 11:654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.