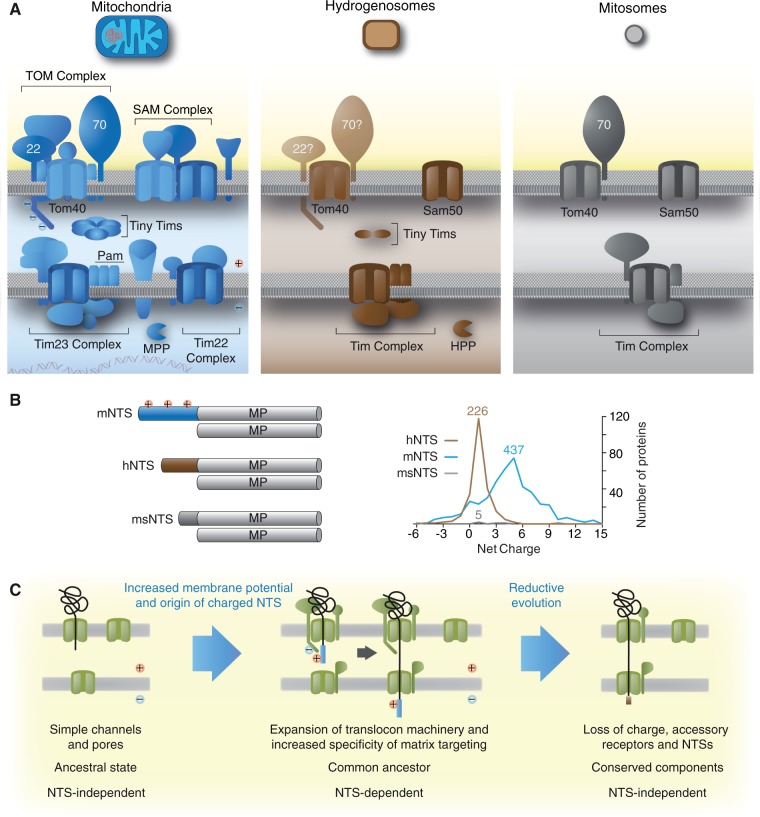
Fig. 5.—
The complexity of the mitochondrial targeting machinery reduces together with the evolutionarily reduction of the organelle’s biochemistry. (A) An illustration of the organellar import machineries from mitochondria, hydrogenosomes, and mitosomes. It is estimated that classical mitochondria import between 1,000 and 1,500 proteins, while hydrogenosomes of Trichomonas import only 200–500 proteins. (B) Mitochondrial matrix proteins carry long NTSs that are usually positively charged, but a few lack a recognizable mNTS. If present, the hydrogenosomal NTS is shorter and no longer charged. Mitosomal proteins of Encephalitozoon cuniculi completely lost the N-terminal targeting information and solely rely on internal motifs. Mitosomes of E. cuniculi are streamlined to such a degree, where they might require to import as little as 22 proteins from the cytosol; proteins that are largely responsible for the last known remaining biochemical pathway, which is iron–sulfur cluster (Fe–S) biosynthesis. (C) Early during mitochondrial evolution, transmembrane protein import was NTS independent. Import complexity (amount of proteins involved and the required targeting information) evolved downstream. During reductive evolution of the organelle, for instance in anaerobic parasites, import complexity decreased back, toward NTS-independent recognition of organellar proteins, translated in the cytosol.