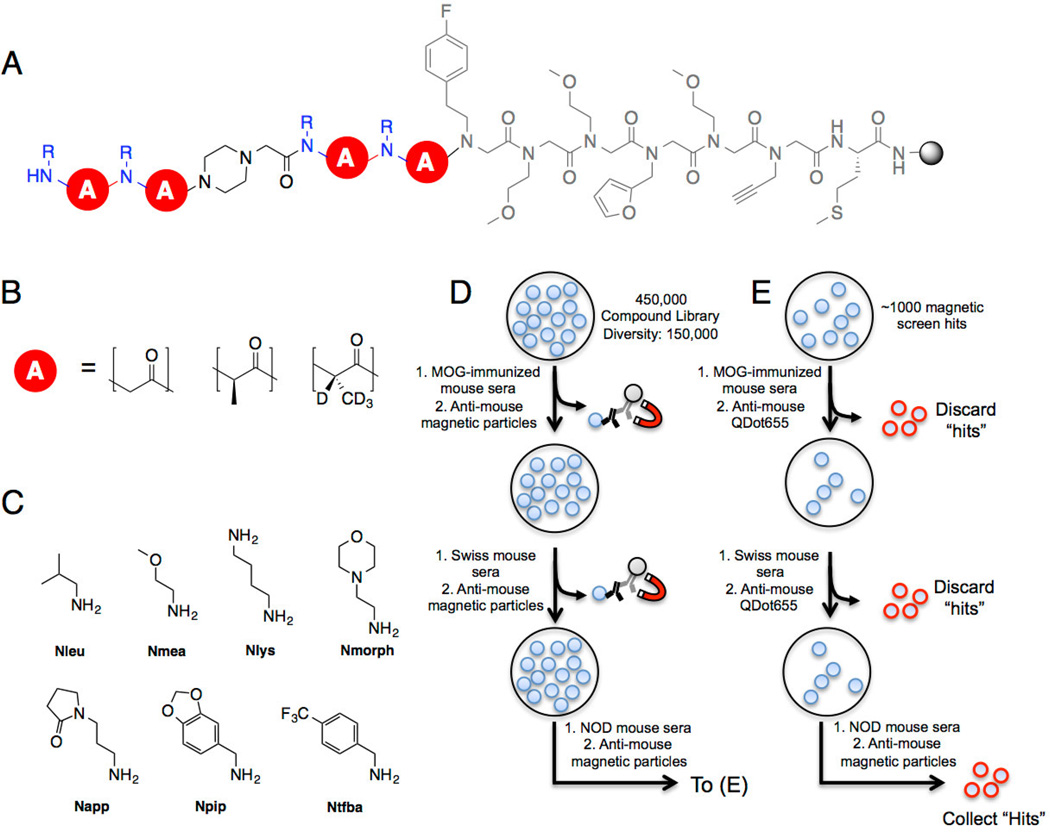
Figure 1. Conformationally constrained PTA-peptoid hybrid library composition and screening.
A) OBOC library design on 90 µm TentaGel macrobeads. The invariant linker is shown in gray. B) The library backbone (red circle) consists of achiral peptoid units or peptide tertiary amide (PTA) units. The R-PTA is deuterated to allow post-screen decoding of stereochemistry by tandem mass spectrometry. C) Seven primary amines were used to install the amide side chains at each position. MMT was used to protect diaminobutane because it is removed under mildly acidic conditions. The total diversity of the library is 150,000 compounds. D) Three copies of the library (450,000 compounds total) were screened against control mouse sera to remove ligands that bound uninteresting IgG or to invariant antibody structural elements. Beads that captured significant antibody were removed via magnetic pull-down after incubation with anti-mouse IgG-coated iron oxide particles. The non-binding beads were incubated with NOD mouse serum in blocking buffer and magnetic anti-mouse IgG and “hit” beads were isolated using a magnetic pull-down. E) The hits isolated from the magnetic screen were stripped of bound protein and incubated with control mouse serum and visualized using red quantum dot-conjugated anti-mouse IgG. Beads that displayed a red halo were removed manually. The remaining beads were incubated with NOD mouse serum and beads displaying a red halo were collected. After cleaving the ligand from each bead, the ligand identity was resolved using tandem MALDI-TOF MS/MS.