Figure 3. Impact of database quality on peptide identifications.
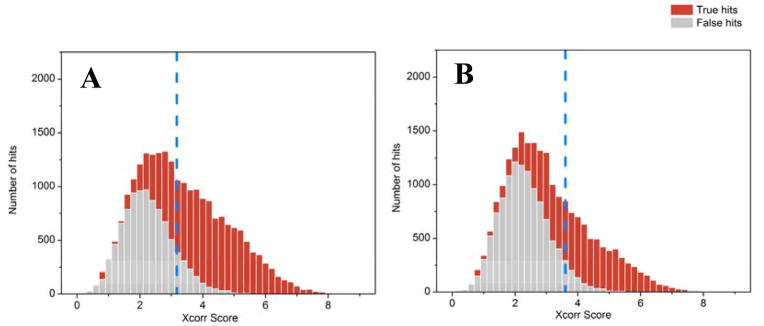
Peptide spectrum matches can be ranked by MyriMatch Xcorr scores to reveal the distribution of true positive (red) vs. false positive (gray) identifications in human adult gut microbiome datasets searched with either a matched metagenome (A) or a pseudo-metagenome assembled from selected microbial isolates (B). An appropriate Xcorr score threshold (indicated by blue dashed line) is chosen to achieve a 1% PSM (peptide spectral match) FDR (false discovery rate; defined by the ratio between false PSMs and total PSMs above the score threshold). The figures reveal that the matched metagenome better differentiates true vs. false distributions, as evidenced by the higher percentage of “red identifications” to the right (i.e. higher Xcorrs) of the dashed line. Even though the pseudo-metagenome likely contains better quality, assembled microbial genomes, the matched metagenome is more closely linked to the actual environmental sample. (Raw data and database details given in reference #70)
