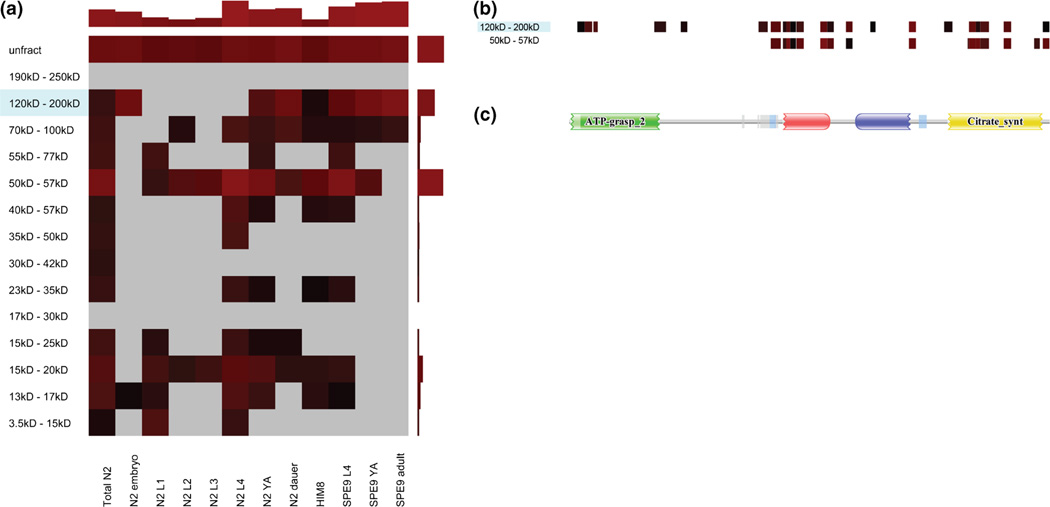
Figure 5.
An example protein heat map and peptide coverage heat map for D1005.1 (a probable ATP-citrate synthase), which may have two possible proteoforms. (a) The protein heat map for the protein D1005.1. D1005.1 has an estimated molecular weight of 121.6 kD, and the gene coding for D1005.1 has no known splice variants (according to WormBase). The NSC for this protein is relatively high in its expected mass fraction (indicated by blue shading); however, it is higher in the 50–57 kD mass fraction. This may indicate some highly sampled peptides that map to D1005.1 also map to another proteoform with a lower mass. Also of note is that no PSMs were found in the lower mass fraction for the embryo developmental stage, whereas a NSC of 108 was calculated for D1005.1 in the higher mass fraction for the same developmental stage. Alternatively, no PSMs were found for D1005.1 in the N2L4 developmental stage in the higher mass fraction, whereas a NSC of 277 was calculated for this stage in the lower mass fraction. This indicates the possibility that each of the two proteoforms is regulated differently with regard to developmental stage. (b) The peptide coverage heat map for D1005.1 for the 50–57 kD and 120–200 kD mass fractions, which shows where the peptides found in the respective mass fractions map to the protein sequence. The lower mass fraction is missing N-terminal peptides found in the higher mass fraction. (c) A domain image generated by PFAM [23] for D1005.1. The missing N-terminal peptides largely correspond to a predicted ATP grasp domain