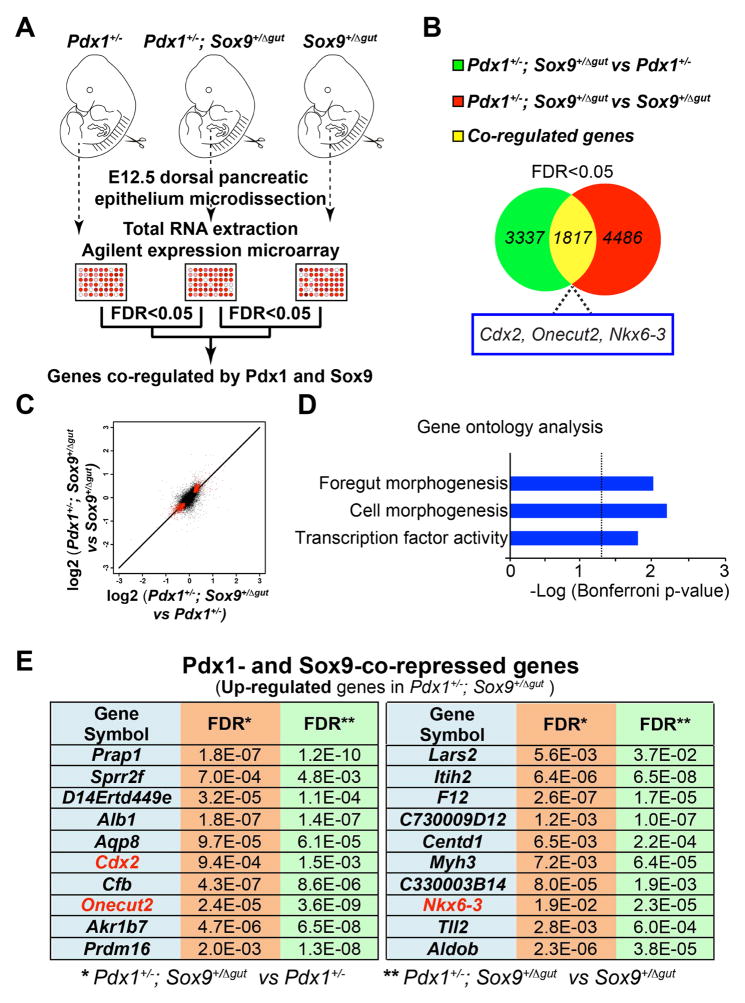
Figure 5. Pdx1 and Sox9 cooperatively silence genes encoding intestinal cell fate regulators.
(A) Illustration of the experimental strategy for gene expression microarray analysis. The mRNA profiles of embryonic day (E) 12.5 pancreata (n=12 per genotype) from 1.) Pdx1+/− versus Pdx1+/−;Sox9+/Δgut and 2.) Sox9+/Δgut versus Pdx1+/−;Sox9+/Δgut littermates were compared. (B) 3,337 and 4,486 genes were differentially-expressed between 1.) and 2.), respectively. A total of 1,817 genes were common to both sets of significantly-regulated genes (FDR<0.05) with the same sign of change (i.e. up-regulated or down-regulated). (C) Pdx1- and Sox9-co-regulated genes were identified by cross-comparing mRNA profiles of E12.5 pancreata (n=12 per genotype) from 1.) Pdx1+/− versus Pdx1+/−;Sox9+/Δgut and 2.) Sox9+/Δgut versus Pdx1+/−;Sox9+/Δgut littermates. A total of 1,817 genes (denoted by red pixels) were common to both sets of significantly-regulated genes (FDR<0.05) with the same sign of change. (D) Gene ontology analysis of the 1,817 Pdx1- and Sox9-co-regulated genes. (E) The top twenty Pdx1- and Sox9-co-repressed genes with the highest fold-change.