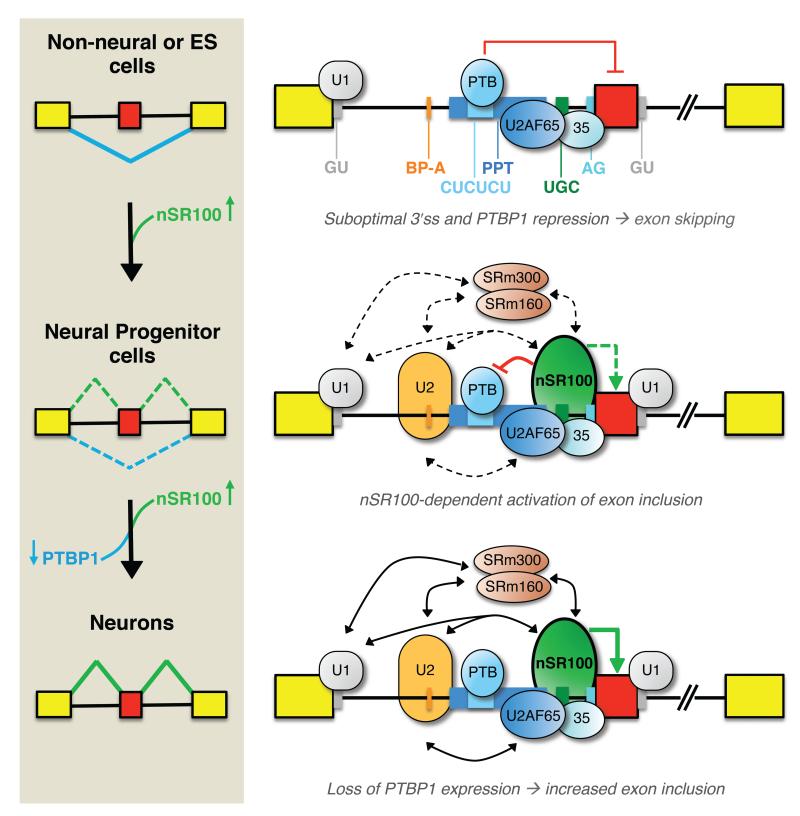
Figure 7. Mechanistic model for nSR100-dependent regulation of neural exon alternative splicing.
Alternative exons in the nSR100-regulated network are associated with a unique arrangement of cis-elements that weaken 3′ splice sites, and act in conjunction with negative regulation mediated by PTBP1 to cause skipping of target exons in non-neural cells. When nSR100 is expressed in differentiating neural precursors and mature neurons it binds to intronic enhancers proximal to 3′ splice sites, and also interacts with multiple early-acting spliceosomal components, to promote exon inclusion. These interactions are sufficient to potently outcompete PTBP1-mediated repression. As neurons develop, PTBP1 is no longer expressed, enabling maximal nSR100-dependent neural exon inclusion.