Fig. 1.
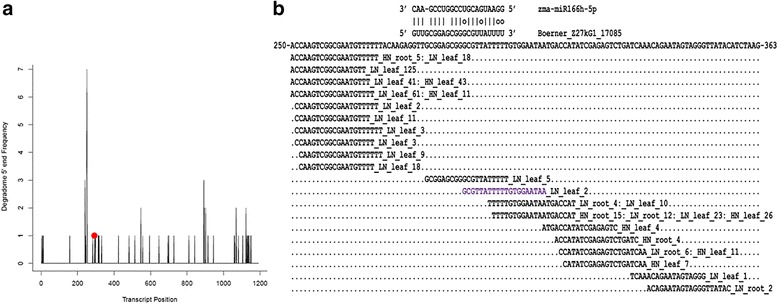
Target plots (t-plots) of the confirmed zma-miR166h-5p target and the distribution of degradome reads among lincRNAs. a Cleavage characteristics of Boerner_Z27kG1_17085, which functions as a zma-miR166h-5p target. The abundance of each sequenced read is plotted as a function of the position of its 5′ end in the transcripts. The peaks of the signatures at the validated cleavage sites of the corresponding miRNAs are shown in red (dots). b miRNA:lincRNA alignment: the reads of the degradome with 5′ ends at the indicated positions are shown in black, while the reads at position 9, 10, 11, or 12 of the inset miRNA target alignment are shown in purple (the cleavage site was counted from the 5′ end of the purple reads, which was position 10 in this example)
