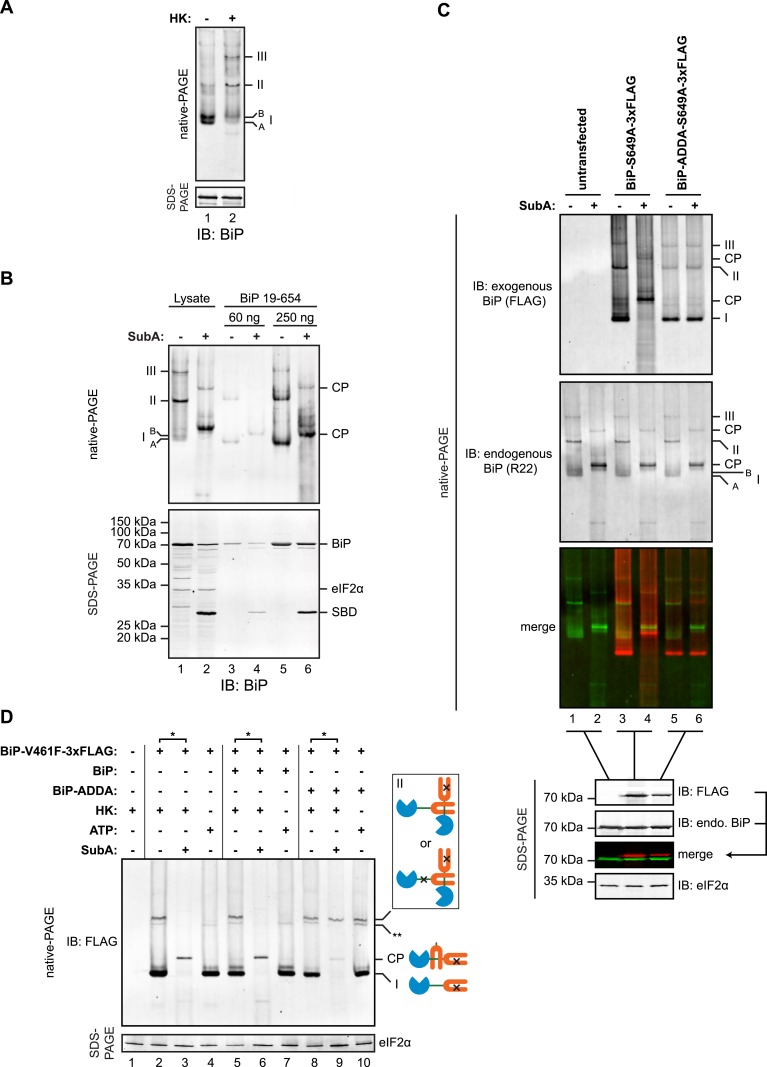
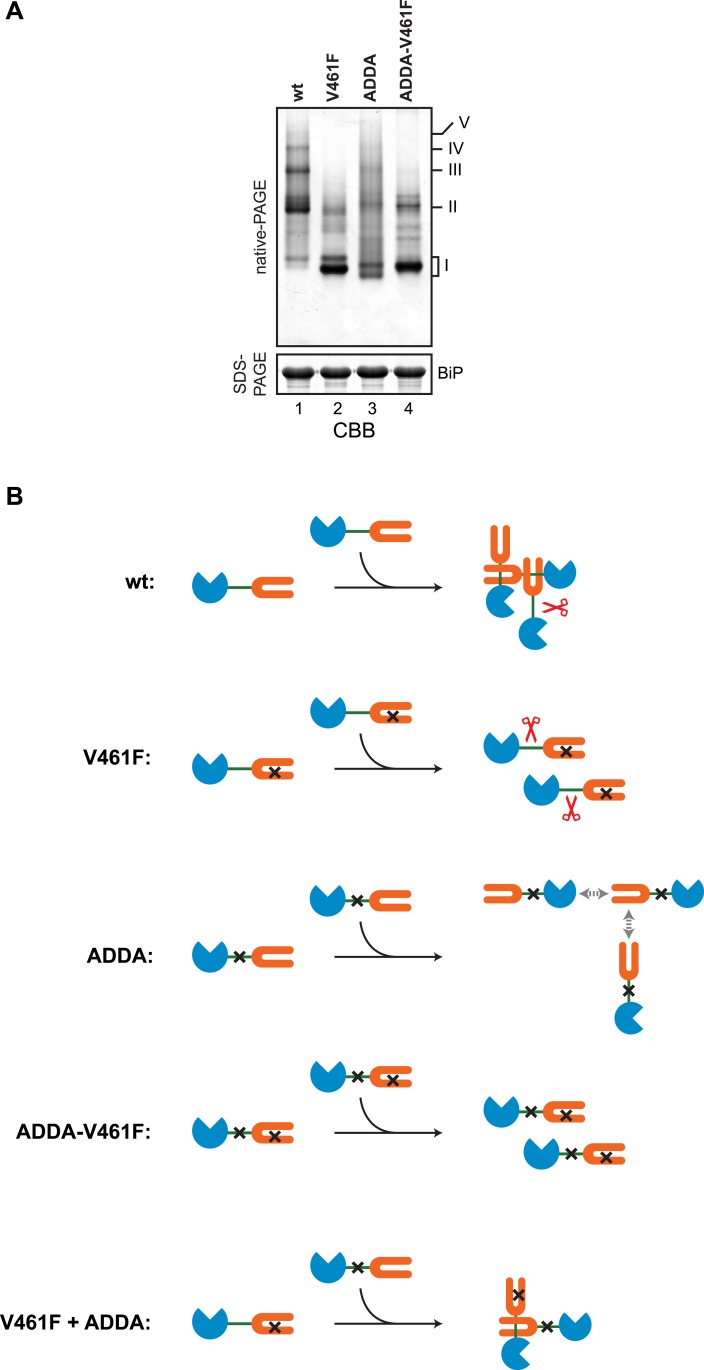
Figure 7. Trans-engagement of the linker by the substrate binding domain (SBD) as the basis for BiP oligomerization in vivo.
(A) Immunoblot of endogenous BiP from lysates of CHO-K1 cells resolved by native-PAGE. Where indicated the lysate was depleted of ATP by incubation with hexokinase and glucose (HK). The major species observed on the native gel have been numbered I-III by order of descending mobility and the two forms of BiP detected in the higher mobility region of the gel (‘A’ and ‘B’) are indicated. Immunoblot of the same sample resolved by SDS-PAGE (lower panel) serves as a loading control. (B) Immunoblot of endogenous BiP from CHO-K1 cells lysed in presence of hexokinase and glucose to deplete ATP (lysate) or mature hamster BiP (19–654) expressed in Escherichia coli and resolved on the same native gel. Where indicated the lysate and or pure BiP were exposed to SubA (5 ng/µl for 10 min at room temperature). The emergent cleavage products (CP) are indicated. Immunoblot of the same sample resolved by SDS-PAGE (lower panel). The free nucleotide binding domain (NBD) is undetected by this antiserum that recognizes the C-terminus of BiP. (C) Anti-FLAG and anti-BiP immunoblots of native gels of ATP-depleted lysates from CHO-K1 cells transfected with the indicated BiP constructs bearing an S649A mutation abolishing their reactivity with the anti-BiP R22 serum. The lysates were split and exposed to SubA as in ‘B’. Anti-FLAG, anti-endogenous BiP (R22) and anti-eIF2α (a loading control) immunoblots of the same samples resolved by SDS-PAGE are presented in the panels below. Note that the FLAG-tagged exogenous BiP proteins are not recognized by the polyclonal anti-rodent BiP antiserum due to the S649A substitution. (D) Anti-FLAG immunoblot of ATP-depleted lysates from CHO-K1 cells transfected with the indicated BiP derivatives and resolved by native-PAGE. Where indicated, the two samples from the same lysate (*) were treated with or without SubA as in ‘B’. ATP (3 mM) was added to lysates loaded in lanes 4, 7 and 10 (prepared without hexokinase and glucose) to dissociate oligomeric species. Anti-eIF2α immunoblot of the same samples resolved by SDS-PAGE is presented in the panel below as a loading control. To aid interpretation, a schema of the BiP species involved is provided to the side of the gel. The BiP NBD is colored blue its SBD orange and the interdomain linker green. The V461F mutation (in the SBD) and the ADDA mutation (in the interdomain linker) are indicated by crosses. Note that the BiP complexes represented by species II in lanes 9 and 10 are resistant to both, SubA and ATP. The identity of the band marked with ** is unknown. Note: To promote BiP oligomerization in vivo, the CHO-K1 cells in panels B-D were treated with thapsigargin (0.5 µM for 10 min before harvest, as will be explained in detail below).