Figure 4.
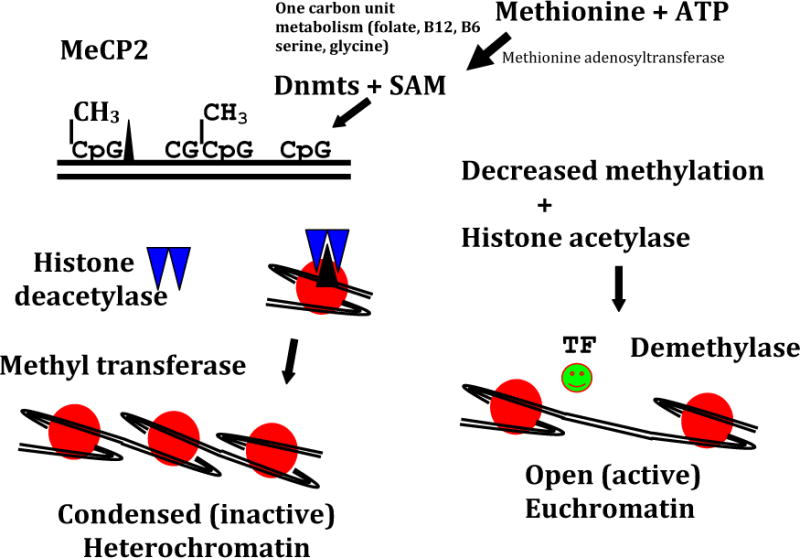
Simplistic scheme showing methylation (CH3) of CpG islands achieved by DNA methyltransferases (Dnmts) and S-adenosyl-L-methionione. Methylated DNA attracts methylated CpG binding protein (MeCP2), which recruits histone deacetylases and histone methylases, resulting in histone deacetylation and methylation. This process occuring in a gene promoter causes heterochromatin formation, resulting in failure of gene expression, which further is stabilized by the binding of the heterochromatin protein (HPI). In contrast, hypomethylation of DNA attracts histone actetylases and demethylases, which results in the oppositie effect with increased euchromatin formation, transcription factor binding, and the activation of gene transcription adenosine triphosphate (ATP). (Reprinted with permission from Devaskar SU, Thamotharan M. Metabolic programming in the pathogenesis of insulin resistance. Rev Endocr Metab Disord 2007; 8:105).
