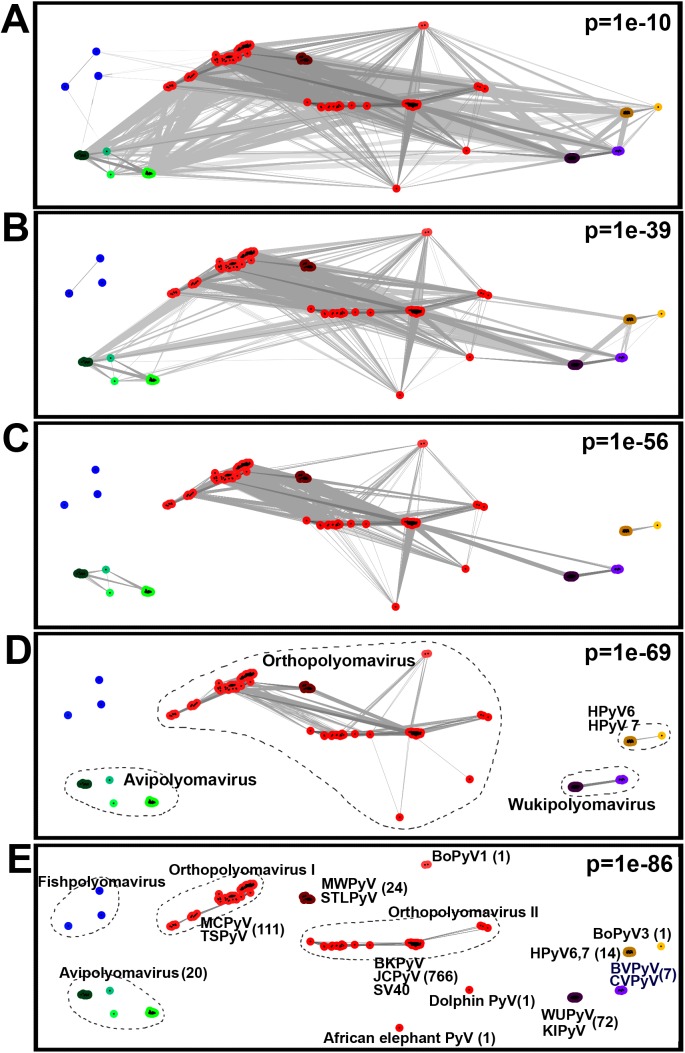
Fig 4. Relationships between PyV genomes, as shown at various cutoff similarity values.
(A) cutoff p = 1x10-10: all PyVs are nested indicating a common nature of all PyVs (not excluding fishpolyomaviruses); (B) cutoff p = 1x10-39: all PyVs are nested, except fishpolyomaviruses; (C) cutoff p = 1x10-56: Avipolyomaviruses are separated; (D) cutoff p = 1x10-69: Wukipolyomaviruses are separated; (E) cutoff p = 1x10-86: Orthopolyomaviruses are divided into 3 main lineages—Orthopolyomavirus I, Orthopolyomavirus II and Malawipolyomavirus. To note, 1x10-86 level indicates closest relationships, while 1x10-10—most distant. Grey lines connect pairs of similar PyV genomes at selected cutoff level of p.