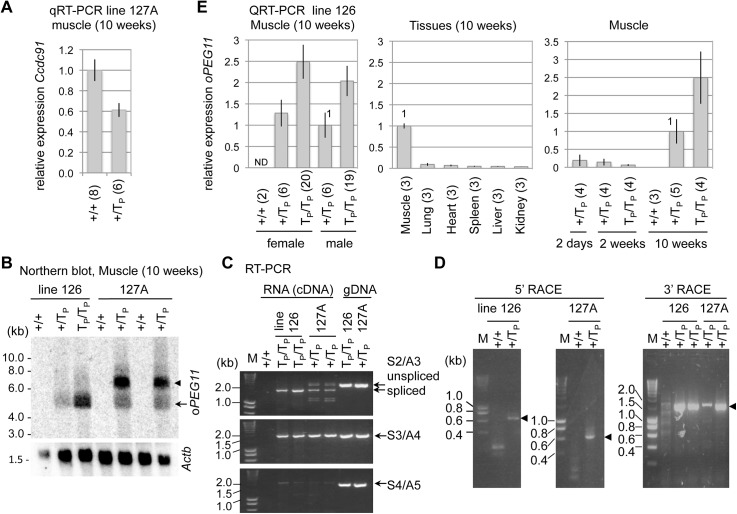
Fig 2. Expression analysis of the oPEG11 transgenes.
A. Relative expression levels of Ccdc91 (mean ± 1.96 x standard error of the mean (s.e.m.)) measured by QRT-PCR using RNA extracted from quadriceps femoris muscles of +/+ and +/TP mice from oPEG11 line 127A. The nearly 1.5-fold reduction in +/TP animals suggests an inactivation of the Ccdc91 gene and likely explains the absence of TP/TP mice in this cross. B. Northern blotting of mRNA extracted from quadriceps femoris of +/+, +/TP, and TP/TP mice from oPEG11 lines 126 and 127A using RNA probes corresponding to the ovine PEG11 transgene (oPEG11) and mouse β–actin for control (Actb). The bands have the approximate size expected for transcripts derived from the oPEG11 transgene (4.3 Kb, arrow) in the both mouse lines plus a larger band (~7 Kb, arrow head) in line 127. C. RT-PCR (or PCR) performed using total RNA (or DNA) extracted from quadriceps femoris (or tail) of +/+, +/TP, and TP/TP mice from lines 126 and 127A using primer combinations shown in Fig 1A. The extra-bands corresponding to unspliced transcripts observed in line 127A are differentiated from the bands expected for spliced transcripts observed in lines 126 and 127A. D. Results of 5’ and 3’ RACE experiments conducted with total RNA extracted from quadriceps femoris of +/+, and +/TP mice from lines 126 and 127A. Sequencing the PCR products marked by the arrows confirmed the use of the vector-specific transcription start and polyadenylation sites (data not shown). E. Relative expression level of oPEG11 (mean ± 1.96 x s.e.m.) in line 126 measured by QRT-PCR using total RNA extracted from quadriceps femoris from animals of indicated genotypes and ages, and from tissues at 10 weeks of age. Expression levels of +/TP animals were used for references. The numbers of tested samples are shown in parentheses.