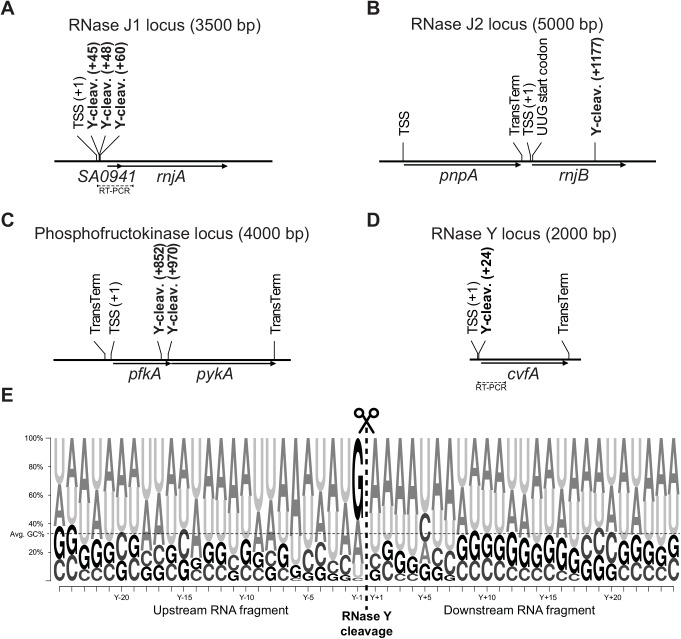
Fig 2. RNase Y cleavage sites fall within or near the genes encoding components of the putative degradosome, as defined by [17], and show sequence preference for guanosines.
Predicted transcription start sites (TSS), detected RNase Y cleavage sites (Y-cleav.) and predicted transcriptional terminators (TransTerm) are indicated. Thin dotted lines show the sequence amplified by RT-PCR to demonstrate that RNase Y cleavage sites and ORFs are potentially on the same RNA molecules. (A) SA0941-rnjA operon, encoding a hypothetical protein and RNase J1. (B) rnjB gene encoding RNase J2 (pnpA encodes PNPase). (C) pfkA-pykA operon, encoding phosphofructokinase and pyruvate kinase. (D) The RNase Y gene (cvfA) with the putative RNase Y cleavage site at position +24. (E) RNase Y cleaves preferentially after a purine. 25 nucleotides on each side of the 99 RNase Y sites (see S2 Table) were extracted from the S. aureus N315 genome, and a frequency plot was generated. The RNase Y cleavage site (thick vertical dotted line), and the position detected by EMOTE (Y+1) are indicated. The preference for a guanosine residue immediately prior to the RNase Y cleavage sites is evident. Additionally, there is an enrichment of A's at position Y–6 and of pyrimidines at position Y+5. Finally, it appears that A's or U's are favoured in the region from 11 nt upstream to 7 nt downstream of the cleavage site, however this could also be due to the over-all nucleotide composition of S. aureus, which has 33% G+C (thin horizontal dotted line).