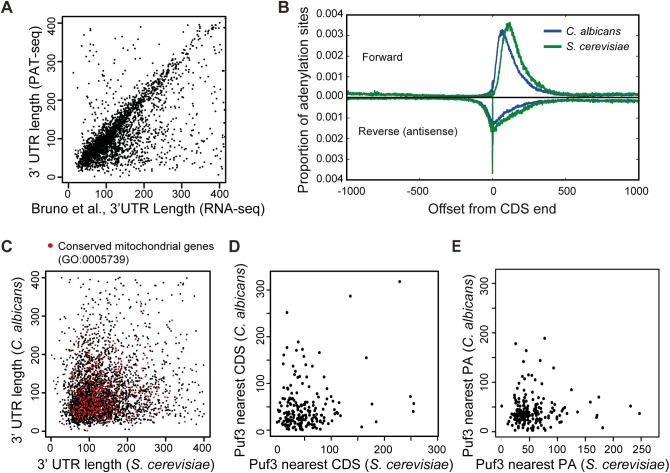
Fig 2. The 3′ UTR landscape of the C. albicans transcriptome.
(A) Comparison of 3′ UTRs as determined by our study and Bruno et al [42]. Of the 3′ UTR that are called by both technologies, 84.5% are within 100 bases (r = 0.4684 (p much < 0.001, n = 2697. Of note, the correlation is highly significant because of the high numbers and would be classed as of moderate strength). Where there is a difference, it is due to filtering differences: we have used the most abundant 3′ UTR, whereas Bruno et al used the longest 3′ UTR for which there was evidence, including minor alternative 3′ UTR isoforms. (B) Graph showing 3′ UTRs are overall shorter in C. albicans than S. cerevisiae. The global positions of polyadenylation in the forward direction and, where it exists, the position of any anti-parallel overlapping adenylated RNA running in the reverse direction. Note any effect of filtering is avoided by this approach as all adenylation sites are utilized in this comparison (370997 and 201547 sites in C. albicans and S. cerevisiae respectively). (C) Comparison between the 3′ UTR lengths of the 3552 orthologous genes between C. albicans and S. cerevisiae. Genes annotated to GO “Mitochondrion” are labeled in red. (D) Comparison of the distance of the Puf3 binding site in putative mRNA targets conserved between S. cerevisiae and C. albicans relative to the closest coding sequence (CDS). (E) Comparison of the distance of the Puf3 binding site in putative mRNA targets conserved between S. cerevisiae and C. albicans relative to the polyadenylation site (PA).