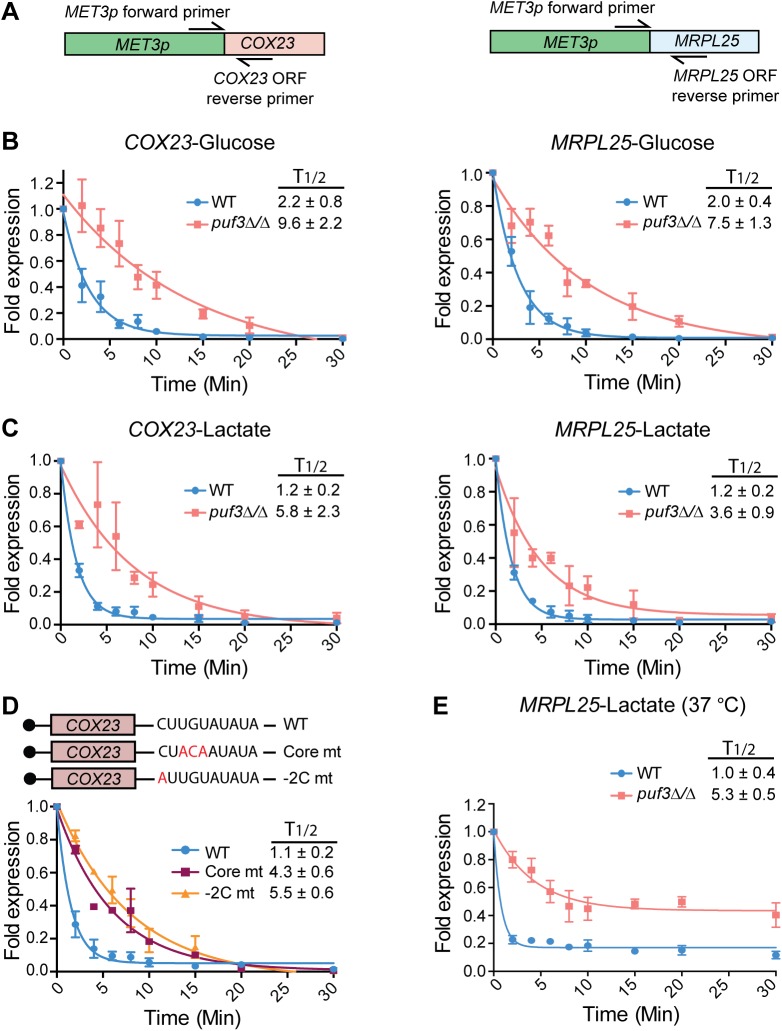
Fig 4. C. albicans Puf3 is a repressor of mRNA stability in glucose and lactate growth conditions.
(A) A cartoon depicting the location of primers used for specific amplification of the MET3p-driven COX23 and MRPL25 genes. Detection of this allele was specific, as demonstrated in S3 Fig. (B) qPCR showing time dependent decay of COX23 and MRPL25 genes following transcriptional shutdown of MET3p in glucose. Fold expression was represented as ratio of expression levels for each time point after dividing with the expression levels at time 0. Decay curves were obtained with the nonlinear regression (curve fit) method using the exponential, one phase decay equation in the GraphPad “Prism 6” software. The half-life (T1/2) was also calculated using this equation by plotting the decay curve of 3–4 biological replicates separately, and is shown as the average ± standard error. (C) The experiments were performed as described in (B), but with lactate as the carbon source. Data represent the average and standard error from 4 biological replicates. (D) The cartoon depicts mutations in the core and -2C positions of the Puf3 binding motif in the COX23 3′ UTR. Decay curves are from wild type C. albicans strains expressing either a COX23 construct with either the wild type or the mutant Puf3 recognition element. The experiments were performed as in (B). Strains were grown in lactate media and shown are results of 2 biological replicates. (E) The experiment was performed as in (C), but concomitant with addition of methionine and cysteine to inhibit transcription, the temperature was raised to 37°C. Shown are results of 3 biological repeats.