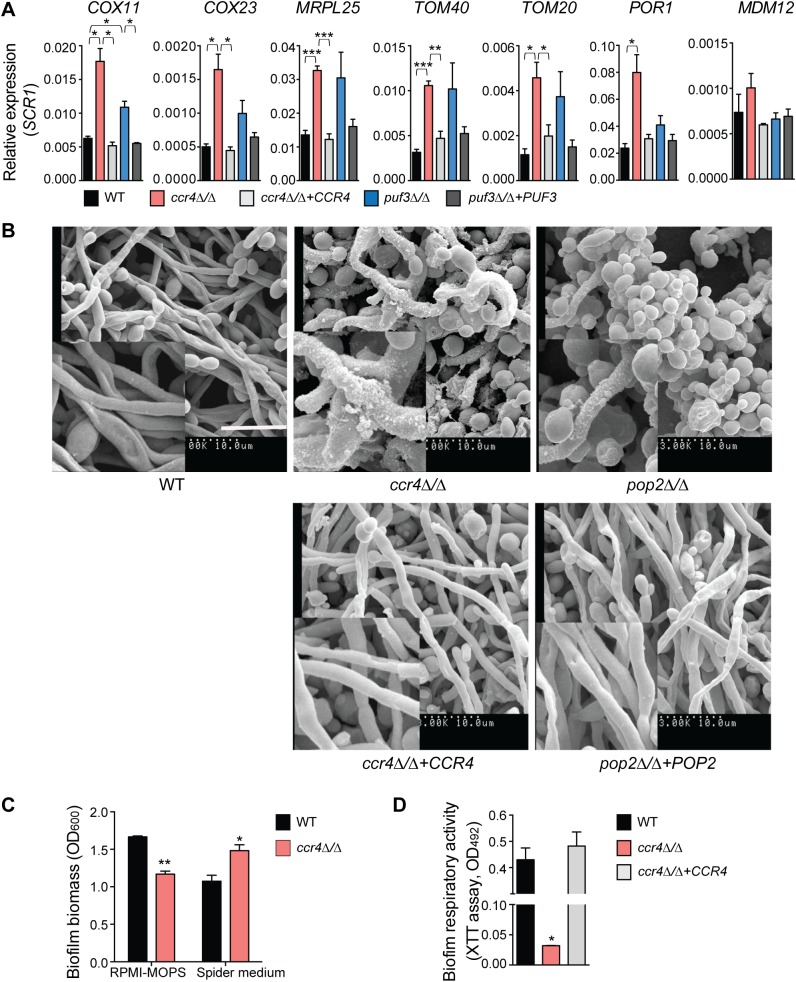
Fig 6. The mRNA deadenylase Ccr4 regulates extracellular matrix production in biofilms.
(A) qPCR analysis of the expression of mitochondrial biogenesis genes in biofilms grown for 48 h in Spider medium. SCR1 RNA was used for normalization. Error bars are ± standard errors of the average of 3 biological replicates. P values are as follows: *** <0.001, ** <0.01, * <0.05. Additional genes are shown in S7 Fig. (B) Scanning electron microscopy of biofilms formed by C. albicans wild type (DAY185), ccr4Δ/Δ and pop2Δ/Δ mutants and their respective complemented strains. Mature biofilms (48 h) grown in Spider medium were assessed. Experiments were repeated at least twice and inset boxes are regional amplifications. Scale bar = 10 μm. Similar results were obtained when biofilms were grown in RPMI-MOPS or YNB (S9 Fig). (C) The data for RPMI-MOPS are from the control samples in the experiments performed to assess biofilm susceptibility to zymolyase (Fig 7B). Biofilms were grown for 48 h in the indicated media. Total biofilm biomass was determined by staining with crystal violet. Bars represent averages ± standard errors from three biological replicates. P value of the difference between WT versus ccr4Δ/Δ is shown as ** <0.01 in RPMI-MOPS and *<0.05 in Spider media. (D) Metabolic activity of C. albicans biofilms was determined using the XTT reduction assay. Results were calculated from three independently grown biofilms for each of the strains assayed in technical duplicates. The error bar represents standard errors. P value of the difference between WT and ccr4Δ/Δ is shown as * <0.05.