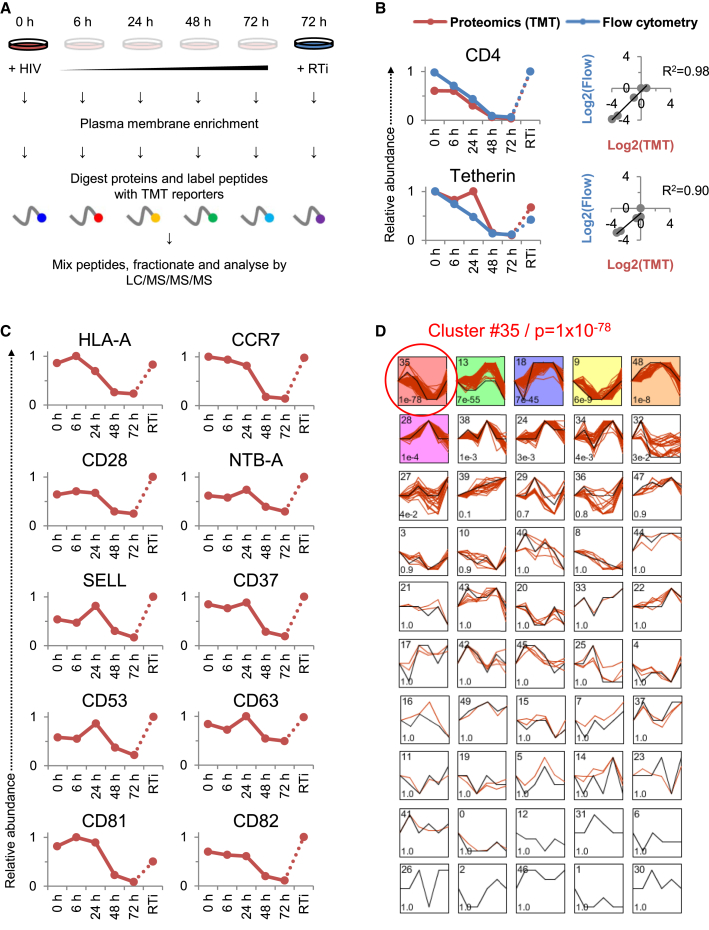
Figure 1.
TMT-Based Proteomic Time Course of Plasma Membrane Protein Expression in HIV-1-Infected Cells
(A) Workflow of TMT-based 6-plex PMP time course experiment. In subsequent figures, time points 1–5 show plasma membrane protein expression 0, 6, 24, 48, and 72 hr after HIV-1 infection (where 0 hr = uninfected cells), and time point 6 shows plasma membrane protein expression 72 hr after HIV-1 infection in the presence of reverse transcriptase inhibitors (RTi). NL4-3-deltaE-EGFP HIV-1 viruses at an MOI of 10 were used for all proteomic experiments.
(B) Comparison of temporal profiles of CD4 and tetherin obtained by proteomic (TMT) versus flow cytometric quantitation. Cells from (A) were stained with anti-CD4 and anti-tetherin antibodies at the indicated time points and analyzed by flow cytometry. Relative abundance is expressed as a fraction of maximum TMT reporter ion or fluorescence intensity. For linear regression, log2(fold change compared with uninfected cells) is shown.
(C) Temporal profiles of previously reported targets for HIV-mediated downregulation.
(D) Identification of enriched temporal profiles by STEM. Model temporal profiles (black) and matched experimental protein expression profiles (red) are shown. Each box includes a profile identification number (top left) and an unadjusted p value (bottom left). Colored boxes indicate model profiles assigned more proteins than expected by chance alone (Bonferroni-adjusted p values < 0.05).