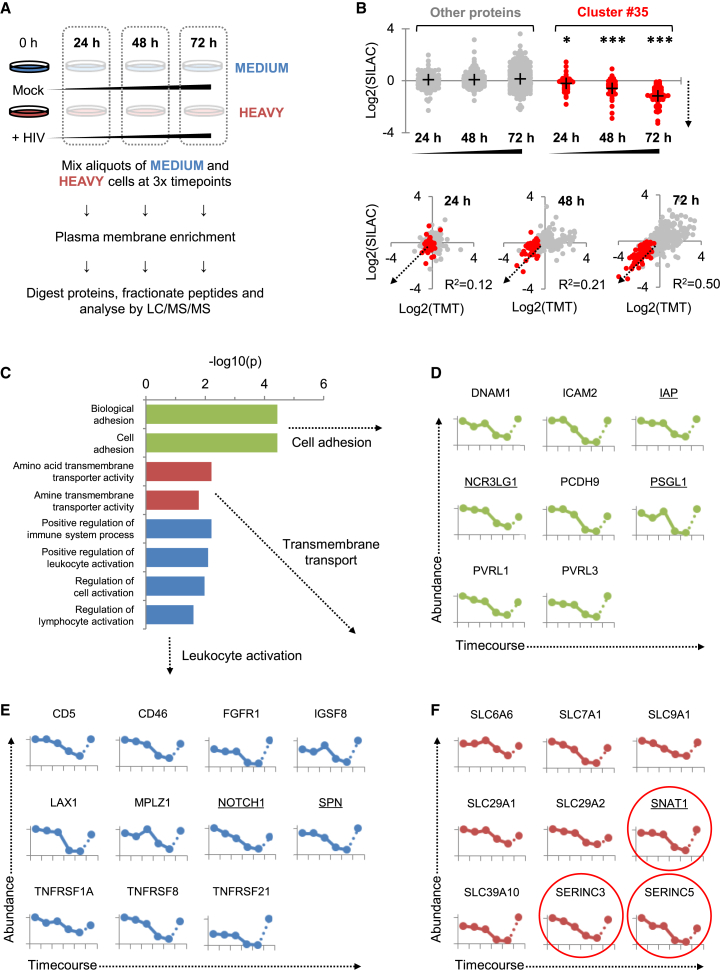
Figure 2.
SILAC-Based Proteomic Validation and Functional Analysis of Cell Surface Targets Downregulated by HIV-1
(A) Workflow of SILAC-based 3-way PMP time course experiment.
(B) Validation of HIV-1 targets (upper panel) and comparison between SILAC- and TMT-based time course experiments (lower panel). Log2(fold change compared with mock/uninfected cells) at 24, 48, and 72 hr is shown for proteins from Cluster #35 (red) versus all other quantitated proteins (gray). Downregulation by HIV-1 is indicated by dotted arrows. Proteins identified by >1 unique peptide in both TMT and SILAC experiments are shown. Crosses indicate mean values. ∗p < 0.05; ∗∗∗p < 0.001.
(C) Gene ontology “molecular function” and “biological process” terms enriched among proteins from Cluster #35. DAVID functional annotation clusters with adjusted p values < 0.05 and containing terms with Bonferroni-adjusted p values < 0.05 are shown. Further details are included in Table S3.
(D–F) Temporal profiles of downregulated proteins associated with cell adhesion (D), leukocyte activation (E), and transmembrane transport (F). Proteomic quantitation and time points are as for Figures 1B–1C. Proteins exhibiting >2-fold downregulation compared with uninfected cells in both TMT and SILAC experiments are shown, and proteins subsequently validated using flow cytometry or immunoblot are underlined.