Fig. 1.
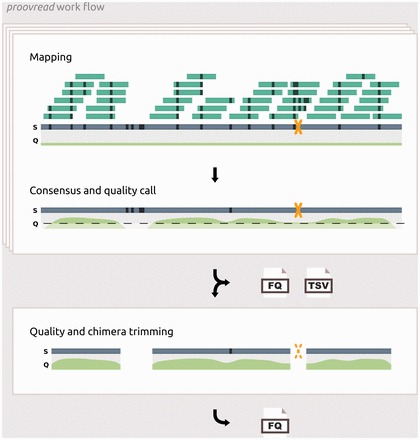
Generic work flow of the proovread correction pipeline. Box 1: Simplified representation of the correction-by-short-read-consensus approach; short reads (dark green bars) are mapped onto an erroneous and chimeric (yellow X) long read (blue bar); black strokes indicate sequencing errors and non-matching alignment positions. Regions with an unusually high amount of errors prevent short read mappings. Base call qualities are represented below the long read (light green curve). During consensus generation, the majority of errors are removed and possible chimeric break points are identified. New qualities scores are inferred from coverage and the composition of the consensus at each position. Processed reads and chimera annotations are written to files. Box 2: The resulting reads are trimmed using a quality cutoff and the chimera annotations generating proovread’s primary output: high-accuracy long reads
