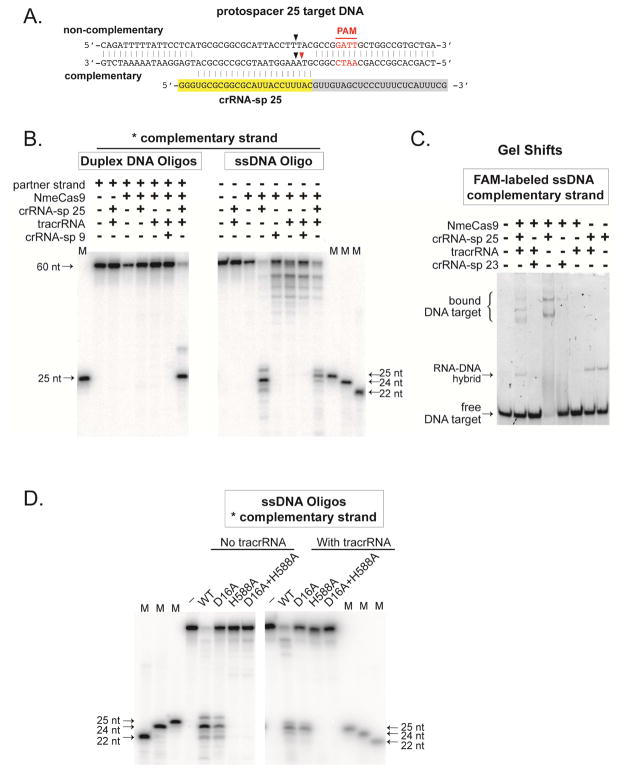
Figure 3. NmeCas9 Exhibits TracrRNA-Independent ssDNA Cleavage Activity.
(A) Schematic representation of ps 25-containing DNA oligonucleotides and the sp 25 crRNA used in (B) and (C). Red lettering, PAM; yellow, crRNA spacer; gray, crRNA repeat; black arrows, predominant cleavage sites for dual-RNA guided NmeCas9 cleavage; red arrow, predominant cleavage site for tracrRNA-independent NmeCas9 cleavage.
(B) NmeCas9 cleaves ssDNA efficiently in a tracrRNA-independent manner. NmeCas9 complexed with small RNAs was assayed for cleavage of double- (left) or single- (right) stranded DNA targets for sp 25. The complementary strand was 5′ 32P radiolabeled. M, size markers. The sizes of substrates, cleavage products and markers are indicated.
(C) NmeCas9 binds an ssDNA target in vitro in a tracrRNA-independent manner. Gel shifts were performed using a 5′ FAM-labeled ssDNA substrate (100nM), NmeCas9 (500nM), and various small RNAs (500nM) as indicated. Binding was performed in cleavage reaction buffer (but with Mg2+ omitted) at room temperature for 10 min, and then resolved by 5% native PAGE.
(D) An intact HNH domain is required for tracrRNA-independent cleavage of complementary strand ssDNA. Wildtype and active-site-mutant NmeCas9s were assayed for cleavage of ssDNA, in the absence (left) or presence (right) of tracrRNA. Reactions were performed as in Fig. 2C. M, size marker. The sizes of substrates, cleavage products and markers are indicated.