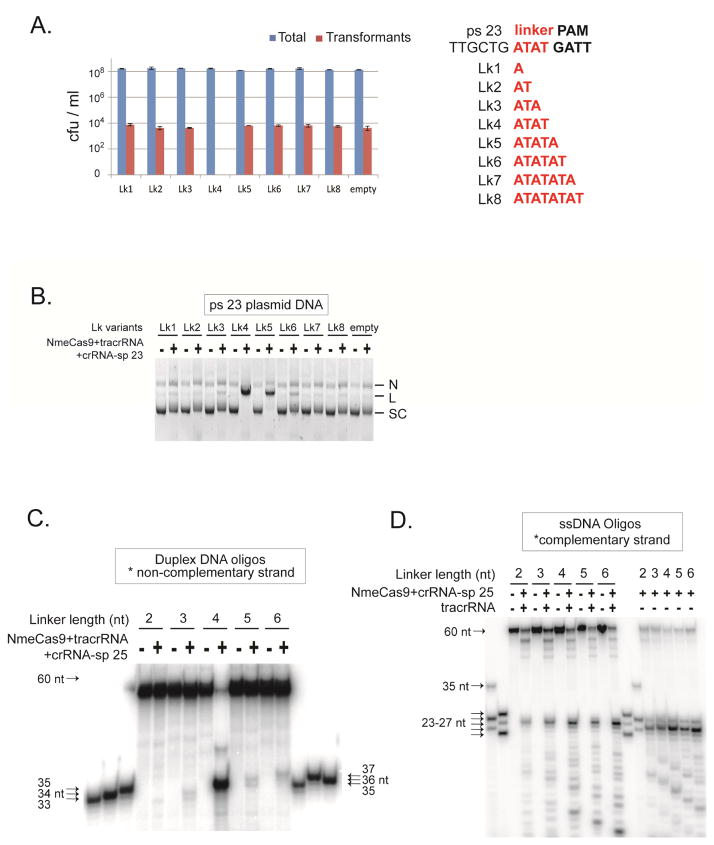
Figure 6. NmeCas9 has Minimal Tolerance for Protospacer-PAM Linker Length Variation.
(A) NmeCas9 only functions with a 4 bp linker in bacterial cells. Left, cellular interference assay. Plasmids containing ps 23 and its linker length mutant derivatives were tested in natural transformation assays as in Fig. 5B. Sequences of the linkers between ps 23 and its PAM in the mutants used in (A) are shown on the right. The linkers are in red.
(B) The same plasmids tested in (A) were assayed for cleavage in vitro by NmeCas9 programmed with sp 23 crRNA and tracrRNA. Reactions were performed as in Fig. 1C. N, nicked; L, linearized; SC: supercoiled.
(C) NmeCas9 cleavage of the non-complementary strand in DNA duplexes is much less efficient when the linker length varies, and the cut site moves in concert with the PAM. The duplex DNAs were 5′ 32P labeled on the non-complementary strand only, and contain ps 25, a flanking GATT PAM and a 2–6 nt linker in between. NmeCas9 was programmed with tracrRNA and sp 25 crRNA. Reactions were performed as in Fig. 2C. M, size markers. The sizes of substrates, cleavage products and markers are indicated.
(D) NmeCas9 cleavage of the complementary strand ssDNA is as efficient when linker length varies, and the cut sites are at a fixed position. The ssDNA targets were 5′ 32P labeled, and contain ps 25, a flanking GATT PAM, and a 2–6 nt linker in between. NmeCas9 was programmed with sp 25 crRNA and with or without tracrRNA, as indicated. Reactions were performed as in Fig. 2C. M, size markers. The sizes of substrates, cleavage products and markers are indicated.