Fig. 4.
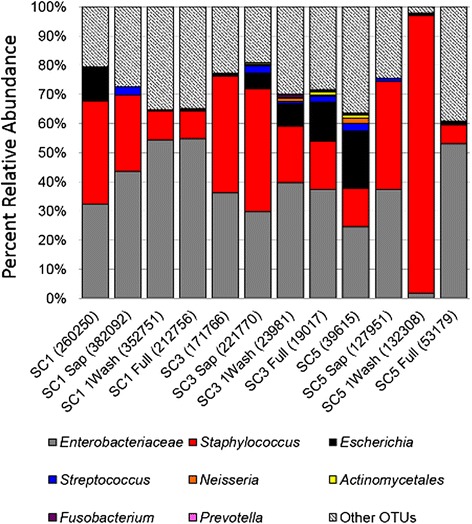
OTU abundance of 16S rRNA Illumina sequenced DNA from synthetic communities. Taxonomic summaries for the synthetic community samples after each step in the saponin blood-treatment protocol were compared. Each bar represents the total PCR amplified DNA sequenced for the sample and the relative abundance of each OTU in the molecular profile. The representative sequence for each OTU was aligned to both the NCBI and HOMD 16S databases in order to determine what synthetic community organism they represented. E. hormaechei sequences were represented by the Enterobacteriaceae OTU, S. aureus was represented by the Staphylococcus OTU, K. pneumoniae by the Klebsiella OTU, E. coli by the Escherichia OTU, S. pneumoniae and S. intermedius by the Streptococcus OTU, N. flava by the Neisseria OTU, M. luteus by the Actinomycetales OTU, F. necrophorum by the Fusobacterium OTU, and P. melaninogenica by the Prevotella OTU. All OTUs with sequence alignments that could not be correlated to the bacteria spiked into the synthetic community were combined into “Other OTUs”, which accounted for 20–40 % of the total OTU abundance
