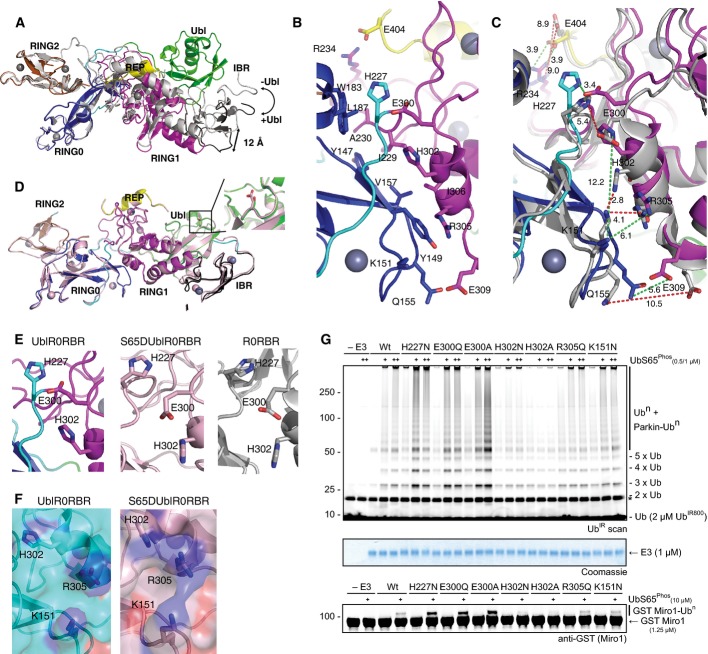
Figure 3.
- Overlay of UblR0RBR (coloured according to Fig1C) and R0RBR (grey) structures. Absence of the Ubl domain causes a hinge opening at the RING0/RING1 interface.
- Close-up of the RING0/RING1 interface in UblR0RBR showing key residues that are involved in ionic/hydrogen bond interactions.
- Overlay of the RING0/RING1 interface from UblR0RBR (coloured) and R0RBR (grey). Green dashes show distances in Å between residues in UblR0RBR, and red dashes show distances between residues in R0RBR.
- Structure of S65DUblR0RBR (pink) overlaid with UblR0RBR parkin (coloured according to Fig1C). The position of D65 or S65 is shown in inset.
- Comparison of the hinge in UblR0RBR (left), S65DUblR0RBR (middle) and R0RBR (right).
- Surface of UblR0RBR (left) and S65DUblR0RBR (right) showing continuous basic patch formed by hinge opening.
- Ubiquitination assays of mutations in the RING0/RING1 interface. Mutations in residues that pin H302 in the flipped-in position are activating, mutation of the basic patch prevents parkin activity, even in the presence of increasing concentrations of phosphoubiquitin (+). This is the case for both chain formation (top) and Miro1 ubiquitination (bottom). A Coomassie-stained loading control is shown in between. A non-specific, ATP-independent band is indicated (*).