Abstract
Diversification of neuron classes is essential for functions of the olfactory system, but the underlying mechanisms that generate individual olfactory neuron types are only beginning to be understood. Here we describe a role of the highly conserved HMG-box transcription factor SOX-2 in postmitotic specification and alternative differentiation of the Caenorhabditis elegansAWC and AWB olfactory neurons. We show that SOX-2 partners with different transcription factors to diversify postmitotic olfactory cell types. SOX-2 functions cooperatively with the OTX/OTD transcription factor CEH-36 to specify an AWC “ground state,” and functions with the LIM homeodomain factor LIM-4 to suppress this ground state and drive an AWB identity instead. Our findings provide novel insights into combinatorial codes that drive terminal differentiation programs in the nervous system and reveal a biological function of the deeply conserved Sox2 protein that goes beyond its well-known role in stem cell biology.
Keywords: C. elegans, neuron differentiation, sox-2, homeodomain
Introduction
The nervous system is composed of an immense variety of neuronal cell types to process information and control behaviors. This is especially important in developing sensory systems, which generate a large number of specialized neurons for sensing various stimuli in the environment. The molecular mechanisms that control neuronal cell type diversification within sensory systems are only partly understood, and there appear to be substantial differences between cell type diversification within distinct sensory systems and across different animal species (Rister et al, 2013).
The Caenorhabditis elegans amphids, a pair of bilaterally symmetric sensory organs on the left and right sides of the head, contain twelve neuron pairs whose functions have been defined by laser ablation: Eleven neuron pairs are chemosensory and at least one neuron pair is thermosensory (Bargmann, 2006). These sensory neurons can be distinguished by their cell body morphologies and positions, axonal trajectories, cilia morphologies, synaptic targets, gene expression patterns, and functions (Lanjuin & Sengupta, 2004). Each sensory neuron type expresses characteristic sensory receptor genes and signaling molecules that serve as molecular markers for its cell identity (Bargmann, 2006). Three pairs of olfactory neurons, AWA, AWB, and AWC, are dedicated to sensing volatile odorants. AWA and AWC neurons mediate attraction behaviors toward specific odors, while AWB neurons mediate repulsion away from particular odors (Bargmann, 2006). Unlike other systems in which each olfactory neuron expresses as few as one olfactory G protein-coupled receptor (GPCR) per neuron, worm olfactory neurons coexpress multiple olfactory receptors, possibly more than 20 per cell (Troemel, 1999).
How does the nematode olfactory system develop? AWA, AWB, and AWC share no common lineage history, and their identity is controlled through distinct regulatory programs employing neuron-type-specific terminal selector transcription factors (Lanjuin & Sengupta, 2004; Hobert, 2010). AWA neurons require the odr-7 nuclear hormone receptor gene for their correct differentiation (Sengupta et al, 1994), AWB neurons are specified by the LIM homeobox gene lim-4 (Sagasti et al, 1999), and AWC neurons are specified by the Otx-type homeobox gene ceh-36 (Lanjuin et al, 2003; Kim et al, 2010). Each of these selector genes initiates and likely maintains the expression of multiple identity features of the respective olfactory neuron, including olfactory receptors.
Intriguingly, loss-of-function mutations in lim-4 lead to a cell identity transformation, such that AWB identity is lost and an AWC identity is adopted instead (Sagasti et al, 1999). Moreover, ectopic expression of lim-4 in AWC is sufficient to transform AWC to AWB neurons. These results suggest that lim-4 acts as a molecular switch to distinguish between AWC and AWB neuron identities (Sagasti et al, 1999). The molecular basis for this switch behavior lies in the cross-regulation of selector genes. In addition to activating the expression of AWB terminal identity genes (such as the putative odorant receptor str-1), lim-4 also acts in AWB to repress the expression of ceh-36, the selector gene of the AWC identity (Kim et al, 2010). Lhx7, the murine homolog of lim-4, plays a similar role in switching alternative neuron identities, as a deletion of this gene causes a subtype of cholinergic interneurons to convert into another subtype of GABAergic interneurons (Lopes et al, 2012).
One key issue that remained unaddressed in previous studies is the problem of specificity. Both the AWB selector gene lim-4 and the AWC selector gene ceh-36 are expressed in a number of non-olfactory neuron types (Sagasti et al, 1999; Lanjuin et al, 2003). By what mechanism is their activity directed toward driving either AWB or AWC identity? We show here that both selector transcription factors operate in the context of a neuron-type-specific combinatorial code that uniquely defines AWB and AWC identities. We find that LIM-4 and CEH-36 each act cooperatively with the HMG-box transcription factor SOX-2 to directly activate terminal differentiation genes to diversify olfactory neuron types. The function of vertebrate Sox2 protein is currently under intense study in the context of stem cell biology. Together with another recent report (Vidal et al, 2015), our paper significantly broadens the perspective of Sox2 function by showing that Sox2 can also have a role in terminal neuronal differentiation, through direct regulation of target genes that define the differentiated state of a neuron.
Results
The AWB olfactory neuron is transformed into the AWCON olfactory neuron in ky707 mutants
The AWC olfactory neuron pair is morphologically symmetric, but differentiates in late embryogenesis into two asymmetric subtypes, AWCON, which expresses the GPCR gene str-2, and AWCOFF, which expresses the GPCR gene srsx-3 (Troemel et al, 1999; Chuang & Bargmann, 2005; Bauer Huang et al, 2007). In wild-type animals, only one of the two AWC neurons becomes the induced AWCON subtype (Fig1Ai and B) (Troemel et al, 1999). The ky707 allele was identified from a forward genetic screen for mutants with ectopic expression of the AWCON cell marker str-2 in either a single or a pair of non-AWC neurons (Fig1Aii, Aiii, and B). ky707 behaved in a recessive manner. The position and morphology of the ectopic AWCON neurons in ky707 mutants were similar to those of AWB olfactory neurons. We found that the AWB neurons in ky707 mutants ectopically expressed a number of additional AWC identity markers, including the GPCR gene srt-28, another AWCON marker (Lesch & Bargmann, 2010), the ceh-36 Otx-homeobox gene, a key AWC identity selector (Lanjuin et al, 2003), and the eat-4/VGLUT gene, a defining feature of glutamatergic identity which is normally expressed in AWC, but not AWB neurons (Figs1B and EV1) (Serrano-Saiz et al, 2013). Furthermore, the guanylyl cyclase gene odr-1, which is normally expressed in both AWC and AWB neurons (L'Etoile & Bargmann, 2000), but with different expressivity (Lanjuin et al, 2003), was expressed in the AWB neurons of ky707 mutants in a manner characteristic of normal AWC expression (FigEV1). The srsx-3 gene, a GPCR-encoding gene asymmetrically expressed in the AWCOFF neuron and the pair of AWB neurons, was lost in AWB in ky707 mutants (Fig1B), suggesting that the AWCON but not AWCOFF identity was adopted by AWB. The ky707 mutation did not affect the expression of identity markers of 17 other sensory neurons and interneurons examined (Fig EV1).
Figure 1.
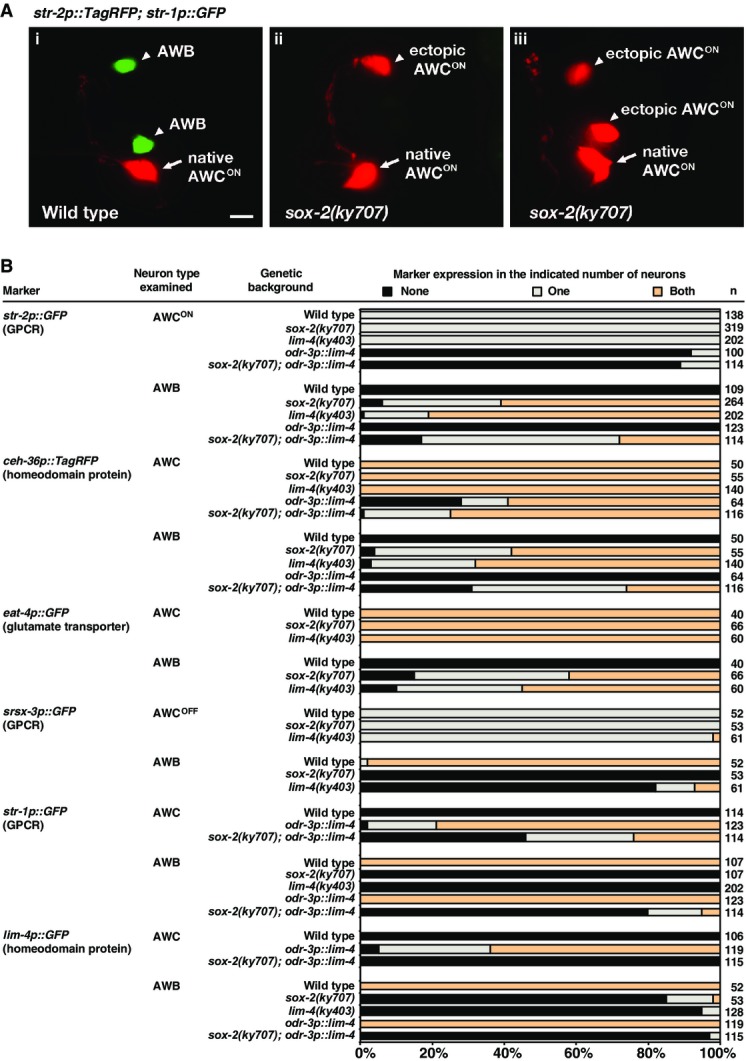
- (Ai) Wild-type animals express str-2p::TagRFP in one AWC neuron and str-1p::GFP in two AWB neurons. (Aii, Aiii) ky707 mutants lose expression of str-1p::GFP in both AWB neurons and gain expression of str-2p::TagRFP in a single AWB neuron (Aii) or in both AWB neurons (Aiii). Anterior is left and ventral is down in ventrolateral views. Scale bar, 5 μm.
- Expression of AWC and AWB markers in ky707 mutants, lim-4 mutants, and odr-3p::lim-4 animals. Animals were scored as adults. n, total number of animals scored. GPCR, G protein-coupled receptor. Expression of additional AWC and AWB markers as well as other neuronal markers in ky707 and lim-4 mutants is included in FigureEV1.
Figure EV1. Expression of additional AWC and AWB markers as well as other neuronal markers in sox-2(ky707) mutants.
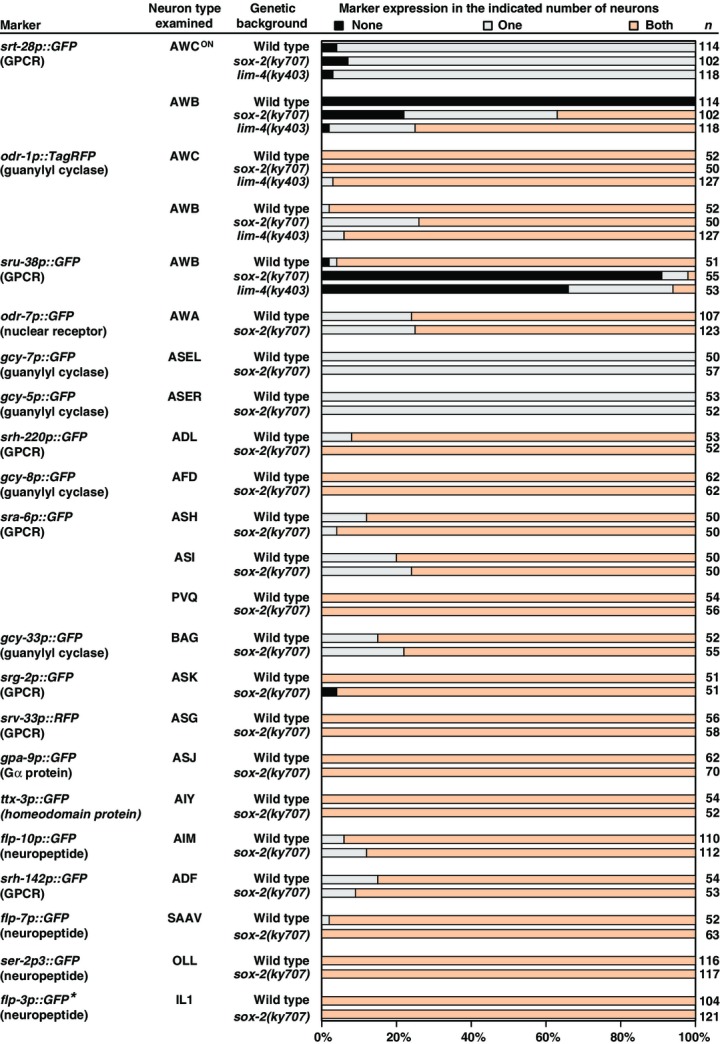
Animals were scored as adults. n, total number of animals scored. *flp-3p::GFP is expressed in 3 pairs of IL1 cells in both wild-type and sox-2(ky707) mutants.
The adoption of the AWCON identity by the AWB neurons in ky707 mutants was paralleled by the loss of the AWB identity. Three AWB identity markers, the str-1 and sru-38 GPCR genes and the lim-4 LIM homeobox gene, failed to be expressed in ky707 mutants (Figs1A and B, and EV1). The apparent identity transformation of AWB to AWC in ky707 mutants is very similar to the transformation observed in animals that lack the lim-4 LIM homeobox gene (Figs1B and EV1) (Sagasti et al, 1999), suggesting that lim-4 and the gene responsible for the ky707 mutant phenotype may work together to distinguish between AWB and AWC neuronal cell identities.
ky707 is a missense mutation in the HMG-box transcription factor gene sox-2
ky707 was mapped to a small interval on the X chromosome, and whole-genome sequencing identified a guanine to adenine transition in the sox-2 gene that results in a glycine (G) to glutamic acid (E) change at the 73rd amino acid of the protein (Fig2A). The AWB-to-AWC transformation phenotype in ky707 mutants was rescued with the transgenes, sox-2p::sox-2 and sox-2ps::sox-2, expressing wild-type sox-2 genomic fragments (Fig2B and C). In contrast, the transgene sox-2p::sox-2G73E, expressing the ky707 mutant form of sox-2 genomic fragment, barely rescued the mutant phenotype (Fig2C). Together, these results support that ky707 is a missense mutation of sox-2.
Figure 2.
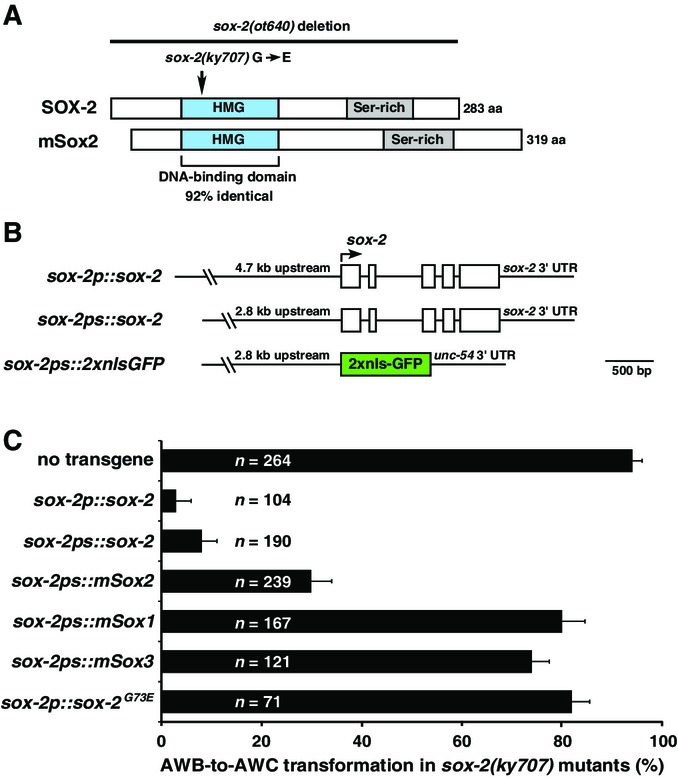
- Domain structure of Caenorhabditis elegansSOX-2 and mouse Sox2 proteins.
- Structure of sox-2 transgenes.
- Rescue analysis of the AWB-to-AWC transformation phenotype with different sox-2 and mouse SoxB1 transgenes in sox-2(ky707) mutants. AWB-to-AWC transformation was determined by the percentage of animals ectopically expressing the AWCON marker str-2p::GFP in one or two AWB neurons. Error bars indicate standard error of proportion.
sox-2 encodes a transcription factor that contains a highly conserved HMG domain at the N-terminus and a diverse serine-rich region at its C-terminus (Fig2A). The HMG domain is a 79-amino acid protein motif that was shown to interact with DNA and/or other proteins (Lefebvre et al, 2007). The C-terminal serine-rich region and the extreme C-terminus of vertebrate Sox2 are essential for transactivation (Ambrosetti et al, 2000). There are five predicted sox genes in C. elegans, eight in Drosophila melanogaster, and 20 in mice and humans (Guth & Wegner, 2008). Caenorhabditis elegans SOX-2 is a SoxB1-type Sox gene with three vertebrate homologs, Sox1, Sox2, and Sox3. The HMG domain of C. elegans SOX-2 protein shares 92% identity to that of human and mouse Sox2 (Figs2A and EV2). The glycine residue affected by the ky707 mutation is conserved in the HMG domain of all SoxB1 proteins (Sox1, Sox2, Sox3) as well as in Sry and Sox14 (FigEV2). However, C. elegans SOX-2 and vertebrate SoxB1 proteins do not exhibit any significant homology outside the HMG domain. To determine which of the three vertebrate SoxB1 genes is most functionally conserved with C. elegans sox-2, we tested whether mouse Sox1, Sox2, and Sox3 under the control of the C. elegans sox-2 promoter are able to rescue sox-2(ky707) mutants. Although the transgenes sox-2ps::mSox1 and sox-2ps::mSox3 showed limited rescue of the ky707 mutant phenotype, sox-2ps::mSox2 displayed the most efficient rescue (Fig2C). This suggests functional conservation between C. elegans SOX-2 and vertebrate Sox2 proteins.
Figure EV2.

Sequence alignment of the HMG domain of Caenorhabditis elegansSOX-2 with HMG domains of human and mouse Sox proteins
Identical amino acids among C. elegansSOX-2, human Sox2, and mouse Sox2 proteins are highlighted in yellow.
Cell identity transformation of AWB into AWCON is partially dependent on the AWC cell identity determination pathway
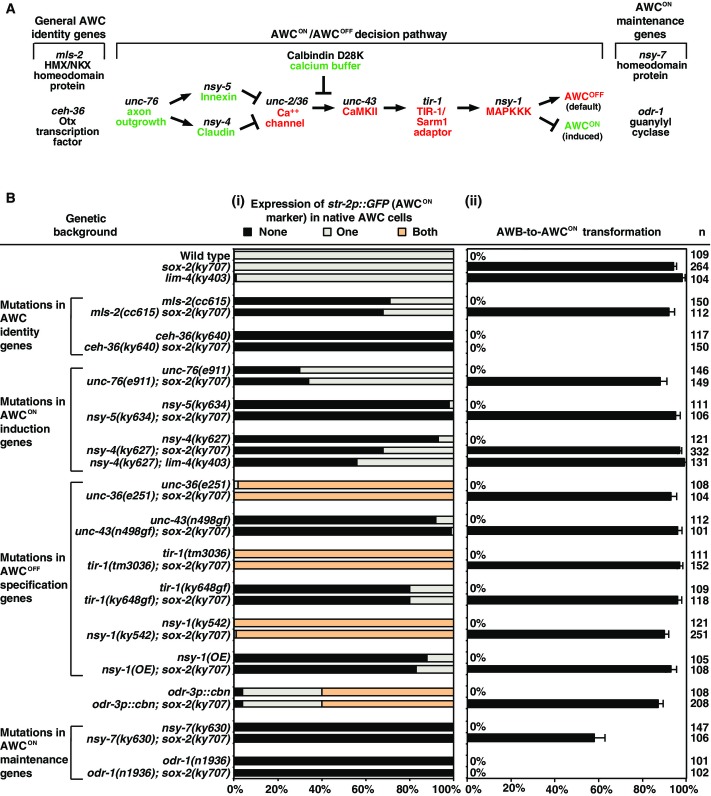
The specification of native AWCON identity is regulated by three developmental events, including the specification of general AWC identity, asymmetric differentiation of the two AWC subtypes (one induced AWCON and one default AWCOFF), and the maintenance of the AWCON subtype (Fig3A and Bi) (Lanjuin et al, 2003; Kim et al, 2010; Taylor et al, 2010; Hsieh et al, 2014). To determine whether the transformation of AWB to AWCON in sox-2(ky707) mutants is dependent on the genes required for the specification of native AWCON identity, we introduced mutations in each of these genes in sox-2(ky707) mutants (Fig3B).
Figure 3.
- The AWC identity determination pathway.
- Expression of the AWCON marker str-2p::GFP in AWC (i) and AWB (ii) neurons in the mutants that are defective in the specification of native AWC identities (Troemel et al, 1999; Sagasti et al, 2001; Tanaka-Hino et al, 2002; Lanjuin et al, 2003; Chuang & Bargmann, 2005; Vanhoven et al, 2006; Bauer Huang et al, 2007; Chuang et al, 2007; Lesch et al, 2009; Kim et al, 2010; Lesch & Bargmann, 2010; Chang et al, 2011; Schumacher et al, 2012). n, total number of animals scored. cbn, calbindin D28K. AWB-to-AWCON transformation was determined by the percentage of animals ectopically expressing the AWCON marker str-2p::GFP in one or two AWB neurons. Error bars represent standard error of proportion.
The Otx transcription factor CEH-36 is a selector of terminal AWC identity, directly controlling the expression of AWC identity features; the HMX/NKX homeodomain protein MLS-2 is a transiently expressed inducer of ceh-36 (Fig3A) (Lanjuin et al, 2003; Kim et al, 2010). The sox-2(ky707) AWB-to-AWCON transformation phenotype was completely suppressed in ceh-36(lf) mutants, but not affected by mls-2, demonstrating that sox-2 cooperates with ceh-36 but normal mls-2 input is not required to turn on ceh-36 expression (Fig3Bii). Mutations in the AWCON maintenance genes nsy-7 (homeodomain-like transcription factor) and odr-1 (guanylyl cyclase) significantly reduced the sox-2(ky707) mutant phenotype (Fig3Bii) (Troemel et al, 1999; Lesch et al, 2009). However, the sox-2(ky707) mutant phenotype was not significantly changed by mutations in any of the AWCON/AWCOFF decision genes (Fig3Bii) (Taylor et al, 2010; Hsieh et al, 2014). Together, these results suggest that transformation of AWB to AWCON in sox-2(ky707) mutants depends on the AWC identity specification genes and the AWCON maintenance genes, but is independent of any of the genes required for AWC asymmetry. These results also indicate that native and ectopic AWCON cells use overlapping but distinct mechanisms to specify and maintain their identities.
The sox-2(ky707) mutation causes functional transformation of AWB to AWCON neurons
AWCON and AWB olfactory neurons have distinct sensory functions: AWCON neurons mediate attraction behaviors toward the odor butanone (Wes & Bargmann, 2001), while AWB neurons mediate repulsion behaviors from the odor 2-nonanone (Troemel et al, 1997). To determine whether AWB is transformed to AWCON at functional levels in sox-2(ky707) mutants, we performed behavioral assays with AWB- and AWC-sensed odors (Fig4).
Figure 4.
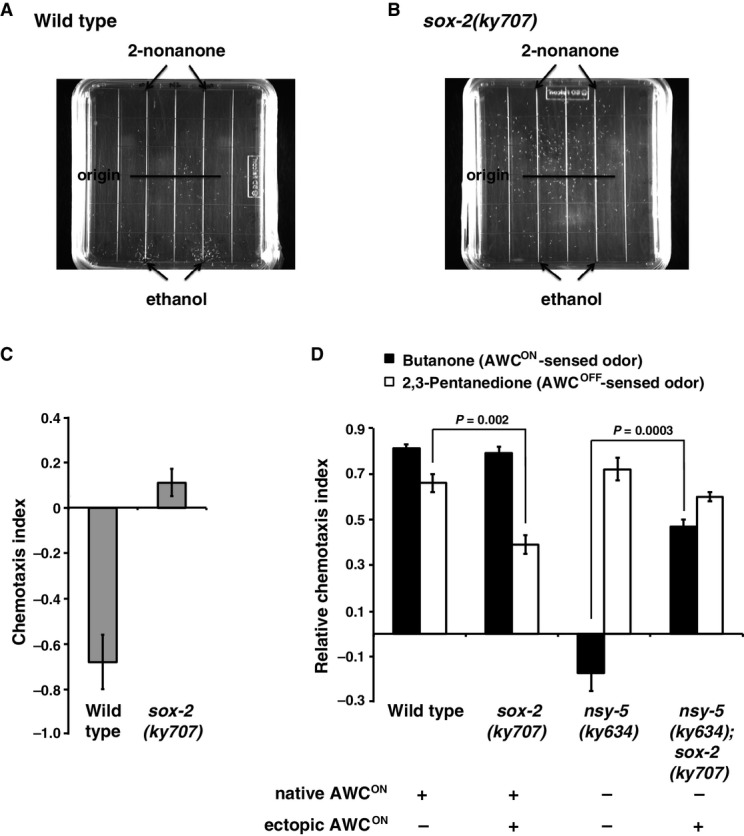
- Chemotaxis assay plate of wild-type worms repulsed by the AWB-sensed odor 2-nonanone. White dots on assay plates are individual worms; black lines on plates represent the location where worms were originally placed on the assay plates.
- sox-2(ky707) mutants lose repulsion to 2-nonanone.
- Quantification of the level of avoidance to 2-nonanone. A more negative number indicates a more repulsive response. Error bars represent standard error of the mean.
- Quantification of chemotaxis indices of worms to butanone and 2,3-pentanedione. A higher index indicates a higher level of attraction. Error bars represent standard error of the mean. Unpaired Student's t-test was used to determine P-value.
In the avoidance assays with 2-nonanone, wild-type worms were severely repulsed by the odor (Fig4A and C). In contrast, sox-2(ky707) mutants lost their ability to avoid the repulsive odor (Fig4B and C), suggesting that ky707 mutants lose AWB neuronal function. In the chemotaxis assays, sox-2(ky707) mutants detected the AWCON-sensed odor butanone normally (Fig4D). However, sox-2(ky707) mutants showed a significantly decreased ability in detecting the AWCOFF-sensed odor 2,3-pentanedione (Fig4D). This result suggests that the sox-2(ky707) mutation may suppress the AWCOFF neuronal function, consistent with the suppression of the nsy-4(lf)2AWCOFF phenotype (in which both AWC neurons become AWCOFF) by sox-2(ky707) (Fig3Bi). nsy-5(ky634) mutants, which fail to induce AWCON cells, lost the ability to detect butanone, but chemotaxed normally to 2,3-pentanedione (Fig4D) (Chuang et al, 2007). nsy-5(ky634); sox-2(ky707) double mutants, which lose native AWCON but acquire ectopic AWCON (Fig3Bi and Bii), were capable of sensing butanone (Fig4D). This indicates that the ectopic AWCON in sox-2(ky707) mutants can function as native AWCON cells. Together, these results suggest a functional transformation of AWB to AWCON neurons in sox-2(ky707) mutants.
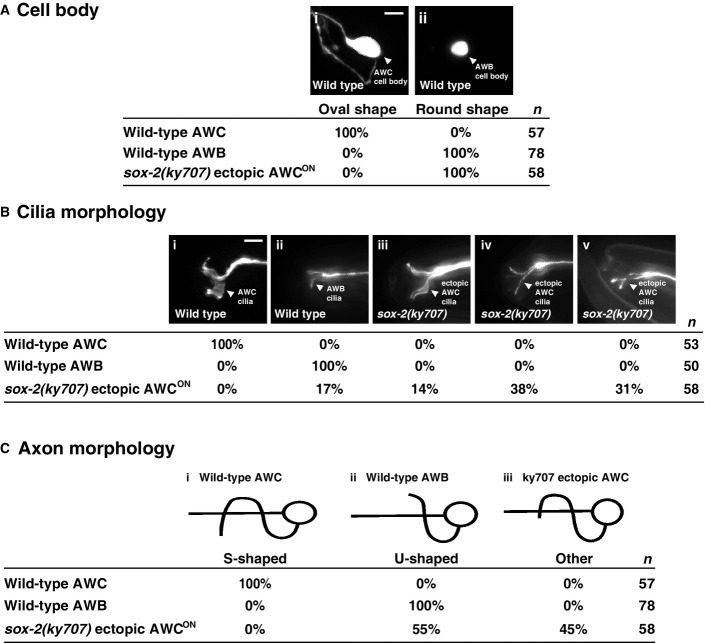
To determine whether ectopic AWC neurons adopted native AWC morphology in sox-2(ky707) mutants, we analyzed cell body, axon, and cilia morphologies of these neurons in wild-type and sox-2(ky707) mutants. The cell body of ectopic AWC neurons in sox-2(ky707) mutants had the same round shape as AWB neurons, different from the characteristic oval shape of native AWC cells (FigEV3A). In addition, the majority of the cilia of ectopic AWC cells displayed variable morphologies more similar to those of wild-type AWB cells than native AWC cells (FigEV3B). These results suggest that the AWB cell body and cilia were not transformed to AWC at the morphological level in sox-2(ky707) mutants. However, 45% of ectopic AWC neurons in sox-2(ky707) mutants had the S-shaped axon morphology similar to that of native AWC neurons (FigEV3C). This result suggests that functional transformation of AWB to AWC neurons in sox-2(ky707) mutants is mainly correlated with the morphological change in axons. This result is also consistent with the important role of AWB and AWC axons in connecting with different sets of sensory neurons and interneurons for mediating distinct behaviors (White et al, 1986).
Figure EV3.
- Wild-type animals have oval-shaped AWC cell bodies (Ai) and small, round AWB cell bodies (Aii). Ectopic AWCON cell bodies are small and round in sox-2(ky707) mutants, similar to native AWB neurons. Scale bar, 5 μm.
- Wild-type AWC cilia have thick, butterfly-shaped morphology (Bi), while AWB cilia are thinner and resemble a tuning fork (Bii). Ectopic AWCON cilia in sox-2(ky707) mutants have either thickened cilia (Biii), prongs that are spread wide apart (Biv), or cilial prongs that appear to cross over each other (Bv). Scale bar, 5 μm.
- AWC axons are S-shaped (Bi), while AWB axons are U-shaped (Bii) in wild-type animals. Ectopic AWCON cells in sox-2(ky707) mutants display native AWC-like axons, which extend beyond the typical AWB U shape but do not continue to form the complete S-shaped morphology of wild-type AWC axons (Biii). To ensure accuracy, axon and cilia morphology of ectopic AWCON was analyzed in the nsy-5(ky634lf); sox-2(ky707) mutants that lost native AWCON and had a single ectopic AWCON neuron.
sox-2 is required to specify both AWB and AWC identities
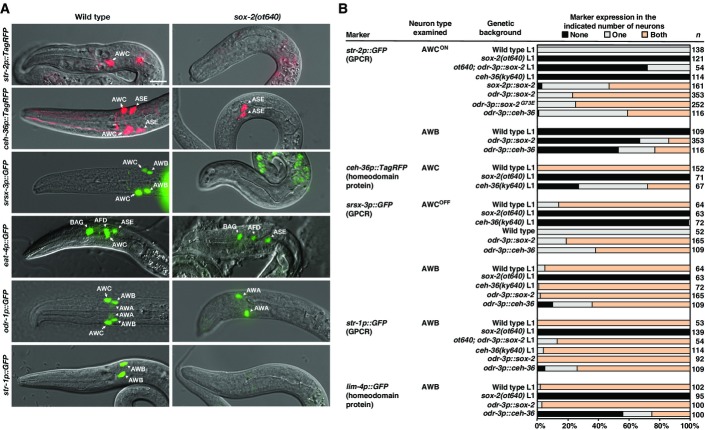
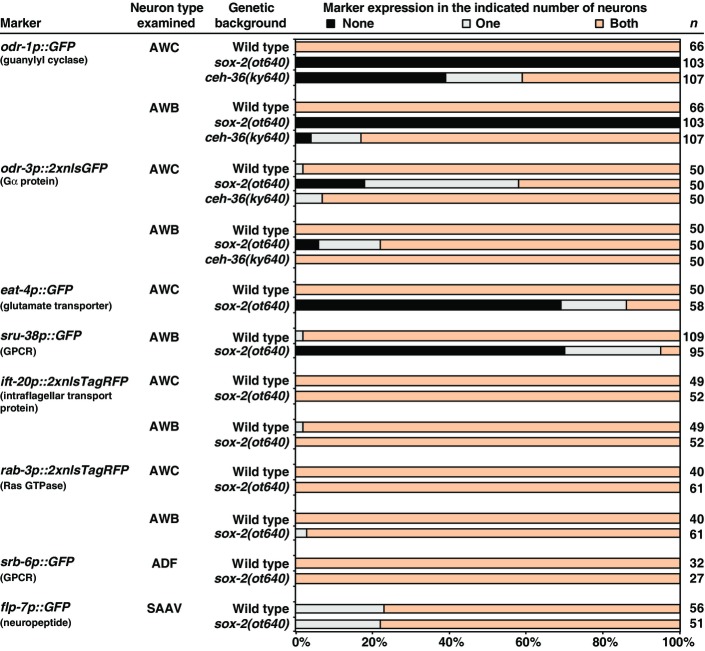
To further dissect the function of sox-2 in AWB and AWC cell identity specification, we examined animals carrying a deletion allele of sox-2, ot640, that eliminates the entire sox-2 coding region (Vidal et al, 2015) and is therefore a null allele. We examined terminal identity markers of AWB and AWC neurons in the sox-2(ot640) null mutants, including AWC-specific genes (str-2, eat-4, and ceh-36), AWB-specific genes (str-1, lim-4, and sru-38), and genes that are expressed in both AWB and AWC neurons (odr-1, odr-3, srsx-3) (Figs5 and EV4). Unlike sox-2(ky707) mutants in which only AWB differentiation was defective, sox-2(ot640) null mutants showed severe terminal differentiation defects of both AWB and AWC neurons (Figs5 and EV4). The only neuron-type-specific identity marker that we found to be unaffected was odr-3 (FigEV4). The expression of the pan-neuronal marker rab-3, a Ras GTPase gene, and the pan-sensory reporter ift-20, an intraflagellar transport gene, was also not affected in AWC and AWB neurons in sox-2(ot640) null mutants (FigEV4). Moreover, the differentiation of the ADF and SAAV neurons (lineal sisters of AWB and AWC, respectively) was not affected in the sox-2 null mutants (FigEV4). These observations indicate that sox-2 controls the terminal differentiation of AWB and AWC, but not their overall generation as neurons, nor does it control earlier lineage decisions. Such differentiation defects are common features of terminal selector transcription factors (Hobert, 2011).
Figure 5.
- Overlay images of DIC and different AWC and AWB markers in wild-type and sox-2(ot640) null mutants. Images were taken at the first larval stage. All sox-2(ot640) animals scored displayed the Pun (Pharynx Unattached) phenotype. Anterior is left and ventral is down. Scale bar, 10 μm.
- Expression of selective AWC and AWB markers in sox-2(ot640),ceh-36(ky640), odr-3p::sox-2, and odr-3p::ceh-36 animals. Animals were scored in the first larval stage (L1) or adults. n, total number of animals scored. Expression of additional AWC and AWB markers as well as ADF and SAAV neuronal markers in sox-2(ot640) and ceh-36(ky640) mutants is included in FigureEV4.
Figure EV4.
Expression of AWC, AWB, pan-neuronal, ADF, and SAAV markers in sox-2(ot640) and ceh-36(ky640lf) mutants
Animals were scored in the first larval stage. n, total number of animals scored.
sox-2 acts cell autonomously in AWC and AWB neurons
To examine the site of sox-2 gene action, we first examined a sox-2 reporter gene. Since 2.8 kb of sequence upstream of the sox-2 start codon, combined with the sox-2 coding sequence, was able to rescue the sox-2 mutant phenotype (Fig2B and C), we fused these 2.8 kb of sequences to 2xnls-GFP. This fusion gene was expressed in the nucleus of both AWC and AWB neurons, identified by an integrated odr-1p::TagRFP transgene (expressed in AWC and AWB) (Figs2B and 6A–C). The expression pattern of sox-2 is consistent with an essential role of sox-2 in the differentiation of AWC and AWB neurons. The expression of sox-2 in AWC and AWB is maintained throughout life. sox-2 is also continuously expressed in other sensory neurons (IL1, IL2, URA, URB, OLL), interneurons (AIM, AIN, AVK, RIH), and motor neurons (RME), as well as in other tissues such as head hypodermis, arcade cells, and rectal epithelial cells (Vidal et al, 2015).
Figure 6.
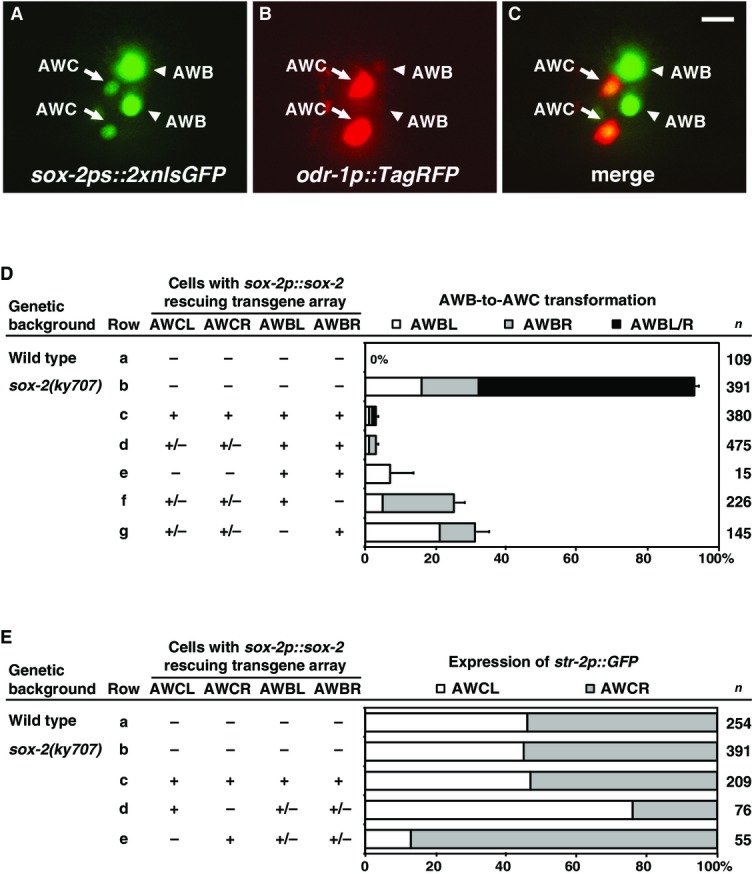
- A–C Expression of sox-2ps::2xnlsGFP (A) and odr-1p::TagRFP (B) in a first-stage larva. Colocalization of GFP and TagRFP in two AWC neurons and two AWB neurons (C). Anterior is left and ventral is down. Scale bar, 5 μm.
- D, E Genetic mosaic analysis of animals expressing the rescuing sox-2p::sox-2; odr-1p::DsRed transgene in ky707 mutants. +, cells that retain the transgene array; −, cells that lost the array. n, number of mosaic animals scored. Error bars indicate standard error of proportion.
To corroborate the notion of cell-autonomous sox-2 gene function, we performed cell-specific rescue experiments and genetic mosaic analysis. When sox-2 was expressed in AWB and AWC neurons with the odr-3 promoter, which is active in the AWB and AWC neurons after they are born (Roayaie et al, 1998), the odr-3p::sox-2 transgene almost completely restored the expression of str-1 in AWB neurons and partially rescued the loss of str-2 expression in AWC neurons in sox-2(ot640) null mutants (Fig5B).
For the mosaic analysis, we utilized animals expressing the transgene sox-2p::sox-2 that rescued the sox-2(ky707) mutant phenotype (Fig6D and E). Caenorhabditis elegans transgenes are maintained as unstable extrachromosomal arrays, which are spontaneously and randomly lost during mitosis. Mosaic animals were identified by expression of the coinjection marker odr-1p::DsRed (expressed in AWC and AWB) that showed which cells retained or lost the rescuing sox-2p::sox-2 transgene. In non-mosaic animals, the sox-2p::sox-2 transgene nearly completely rescued the AWB-to-AWC transformation phenotype (Fig6D, rows a–c). Similar to non-mosaic animals, the majority of mosaic animals that retained the sox-2p::sox-2 transgene in both AWB cells, regardless of the presence of the transgene in both AWCs, showed efficient rescue of the sox-2(ky707) mutant phenotype (Fig6D, rows d and e). When the sox-2p::sox-2 transgene was retained in only one of the two AWB cells, only AWB left (AWBL) or AWB right (AWBR) was transformed to AWC, independent of expression of the transgene in AWC cells (Fig6D, rows f and g). In more than 74% of these mosaic animals that had one AWB transformed to AWC, the sox-2p::sox-2-containing AWB neuron was rescued and the sox-2(ky707) mutant AWB was transformed to AWC (Fig6D, rows f and g). Together, these results are consistent with a cell-autonomous requirement for sox-2 in differentiating AWB from AWC identities.
We also examined the effect of the sox-2p::sox-2 transgene on the AWCON/AWCOFF decision using mosaic analysis (Fig6E). Similar to wild-type, sox-2(ky707) mutants or sox-2p::sox-2 non-mosaic animals did not show a side bias of AWCON induction (Fig6E, rows a–c). When the transgene was retained in only one of the two AWC cells, the sox-2p::sox-2-containing AWC neuron became AWCON and the sox-2(ky707) AWC neuron became AWCOFF over 81% of the time (Fig6E, rows d and e), suggesting that sox-2 has a cell-autonomous ability to promote AWCON.
SOX-2 functions cooperatively with CEH-36 and LIM-4 to diversify olfactory neuron types
As described above, the AWB differentiation defects of sox-2(ky707) mutants displayed striking similarities to the AWB differentiation defects of animals that lack the lim-4 LIM homeobox gene (Figs1B and EV1). Similarly, the differentiation defects of the AWC neurons in sox-2 null mutants phenocopied the defects of loss of the ceh-36 Otx-type homeobox gene in terminally differentiating AWC identity: The expression of eat-4, str-2,srsx-3, and odr-1 was lost in AWC neurons in ceh-36 mutants (Figs5B and EV4) (Lanjuin et al, 2003; Serrano-Saiz et al, 2013), as in sox-2(ot640) null mutants (Figs5 and EV4). Taken together, our phenotypic and genetic analysis led us to hypothesize that sox-2 and ceh-36 may act together to specify the AWC cell identity and that sox-2 may function together with lim-4 to differentiate AWB from AWC identity. In addition, the glycine (G) to glutamic acid (E) substitution at the 73rd residue of the SOX-2 HMG domain in ky707 mutants may specifically affect the ability of SOX-2 to cooperate with LIM-4, but not CEH-36 (Fig1B). Protein–protein interactions involving the HMG domain have been previously described (Lefebvre et al, 2007; Stros et al, 2007).
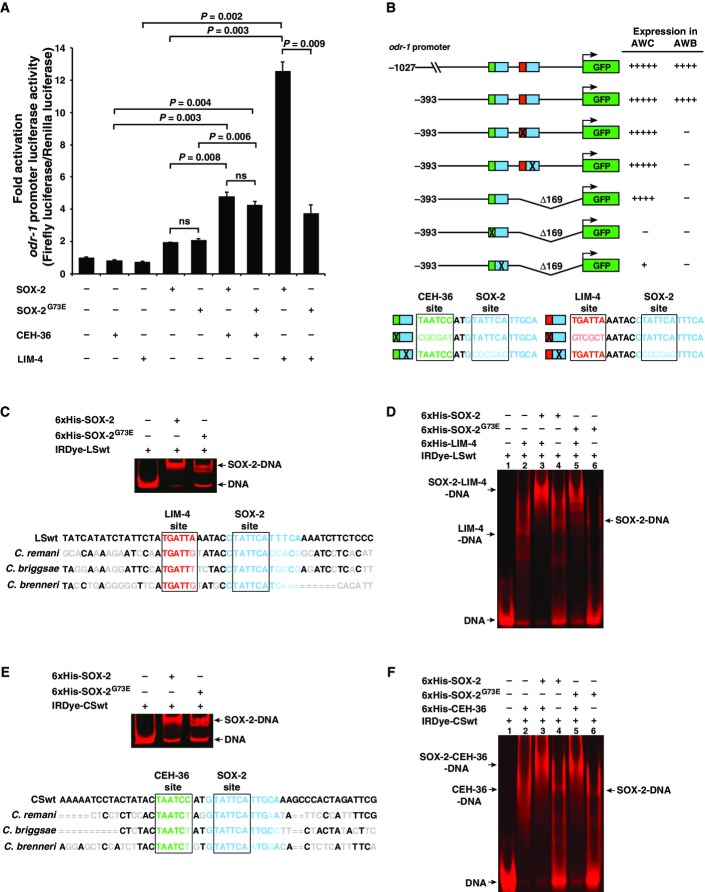
Mammalian Sox2 has been shown to cooperate with different partner proteins in a variety of cell specification processes (Kamachi & Kondoh, 2013). To directly survey the interactions of SOX-2 with CEH-36 and LIM-4, we expressed wild-type SOX-2, mutant SOX-2G73E, CEH-36, and LIM-4 transcription factors in different combinations in COS-7 cells. The promoter of the AWB/AWC marker odr-1 (1,027 bp) was used to express the reporter gene luciferase. When transfected alone, each transcription factor had low activity in activating the odr-1 promoter (Fig7A). In the cotransfection experiments, we found that CEH-36 and LIM-4 had a cooperative effect on the ability of SOX-2 to activate the odr-1 promoter (Fig7A). CEH-36 had a similar cooperative effect with SOX-2 and with SOX-2G73E. However, the cooperative effect of LIM-4 on SOX-2G73E was significantly reduced compared with the effect on SOX-2 (Fig7A). Since SOX-2G73E cooperated normally with CEH-36, the impaired cooperation with LIM-4 rules out that the SOX-2G73E is less stable or less expressed. These results are consistent with a model in which wild-type SOX-2 functions cooperatively with both CEH-36 and LIM-4, while SOX-2G73E can still function cooperatively with CEH-36 but not with LIM-4 (Fig8B). This observation provides a mechanistic basis for the observation that the ky707 allele only affected AWB, but not AWC differentiation. Apart from demonstrating the cooperativity of SOX-2, LIM-4, and CEH-36, the transfection results also demonstrate that these factors operate directly on terminal effectors.
Figure 7.
- A Quantification of fold activation of odr-1 promoter measured using dual-spectral luciferase assays in COS-7 cells. The 1,027-bp odr-1 promoter, expressed in both AWB and AWC neurons, has basal activity in the presence of the single transcription factors CEH-36, LIM-4, SOX-2, or SOX-2G73E. The activity is boosted significantly in the presence of both CEH-36/SOX-2 and LIM-4/SOX-2 combinations. The sox-2(ky707) mutation disrupts the cooperation between LIM-4 and SOX-2G73E, but does not affect CEH-36/SOX-2G73E cooperation. All transfection assays were performed in triplicate and repeated three times; three repeats showed similar trends of results. Data of one set of triplicate are presented. Unpaired Student's t-test was used to determine P-value; ns, not significant.
- B odr-1 promoter GFP reporter constructs and their expression levels in AWB and AWC cells. Increased number of (+) indicates higher intensity of GFP expression; (−) indicates lack of expression. Consensus binding sites of CEH-36, SOX-2, and LIM-4 are boxed. Green, CEH-36 site; blue, SOX-2 site; red, LIM-4 site. Lighter shades of green, blue, and red as well as X represent mutated sites.
- C–F EMSA assays of 6×His-tagged proteins with an IRDye-labeled DNA probe containing LIM-4/SOX-2 adjacent binding sites (CSwt) (C, D) or CEH-36/SOX-2 adjacent binding sites (LSwt) (E, F). Nucleotides in gray, light green, light blue, and light red are sequences of C. remani,C. briggsae, and C. brenneri that differ from C. elegans sequence. = indicates no alignment available between different species. Sequence alignment between species was performed on http://genome.ucsc.edu. Gel images in (D) and (F) were spliced between lanes 2–3 and 4–5 to exclude irrelevant lanes.
Figure 8.
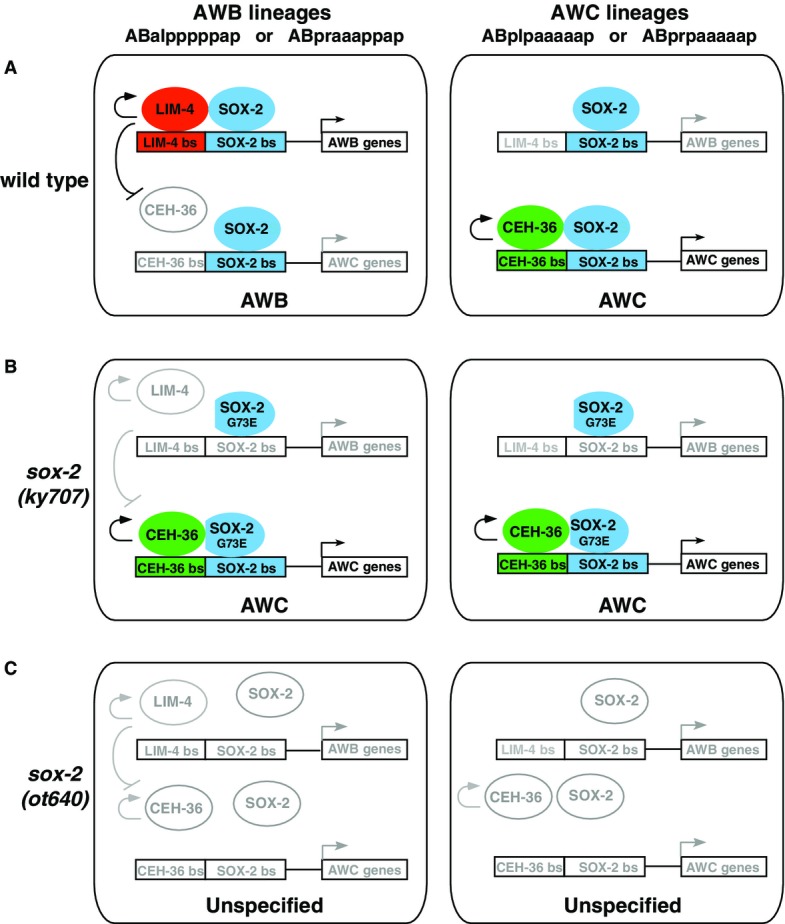
- In wild-type, CEH-36 and SOX-2 cooperate to promote AWC identity in AWC precursor cells (right). In AWB precursor cells, SOX-2 cooperates preferentially with LIM-4 to promote AWB identity, thereby preventing CEH-36/SOX-2 to autoregulate and induce AWC identity (left).
- The sox-2(ky707) mutation has no effect on the binding of SOX-2G73E to the SOX-2 site in the CEH-36/SOX-2 element, allowing CEH-36/SOX-2G73E cooperation to specify AWC identity in AWC precursor cells (right). The sox-2(ky707) mutation specifically reduces the binding of SOX-2G73E to the SOX-2 site in the LIM-4/SOX-2 element, resulting in decreased LIM-4/SOX-2G73E cooperation and consequently enabling CEH-36/SOX-2G73E cooperation to drive AWC identity in AWB precursor cells (left).
- The sox-2 null allele removes all aspects of sox-2 activity, resulting in the loss of lim-4 and ceh-36 activity and, consequently, AWB and AWC identities.
There are several examples of interacting transcription factors that function by cooperative binding to adjacent target sites (Kazemian et al, 2013). Therefore, we searched the odr-1 promoter for putative CEH-36 and LIM-4 binding sites adjacent to SOX-2 consensus sites [SOX-2 sites (Salmon-Divon et al, 2010; Engelen et al, 2011; Fang et al, 2011), LIM-4 sites (Oda-Ishii et al, 2005; Flandin et al, 2011), CEH-36 sites (Kim et al, 2010)]. We identified both predicted LIM-4/SOX-2 and CEH-36/SOX-2 adjacent sites, 2 and 5 base pairs apart, respectively (Fig7B). The LIM-4/SOX-2 and CEH-36/SOX-2 sites are conserved between C. elegans and closely related species C. remanei,C. briggsae, and C. brenneri (Fig7C and E), suggesting that these sites are potentially important cis-regulatory elements. Our promoter-dissecting experiments using GFP reporter transgenes showed that a 393-bp odr-1 promoter region retaining both LIM-4/SOX-2 and CEH-36/SOX-2 sites was sufficient to drive GFP expression, at the same levels as the reporter driven by a 1,027-bp odr-1 promoter, in both AWB and AWC neurons (Figs7B and EV5). In addition, deleting the LIM-4/SOX-2 adjacent sites or mutating either LIM-4 or SOX-2 site in the LIM-4/SOX-2 element in the 393-bp odr-1 promoter region eliminated the expression of reporter transgenes in AWB without affecting the expression in AWC (Figs7B and EV5). This suggests that the LIM-4/SOX-2 sites are necessary for odr-1 expression in AWB. Furthermore, mutating the CEH-36 or SOX-2 binding site in the CEH-36/SOX-2 element caused complete loss or severe reduction of odr-1 expression in AWC, respectively (Figs7 and EV5). This indicates that the CEH-36/SOX-2 adjacent sites are essential for expression of odr-1 in AWC. Together, our results are consistent with our genetic data and also support our model that the predicted LIM-4/SOX-2 and CEH-36/SOX-2 sites are essential for odr-1 expression in AWB and AWC, respectively (Fig8).
Figure EV5.
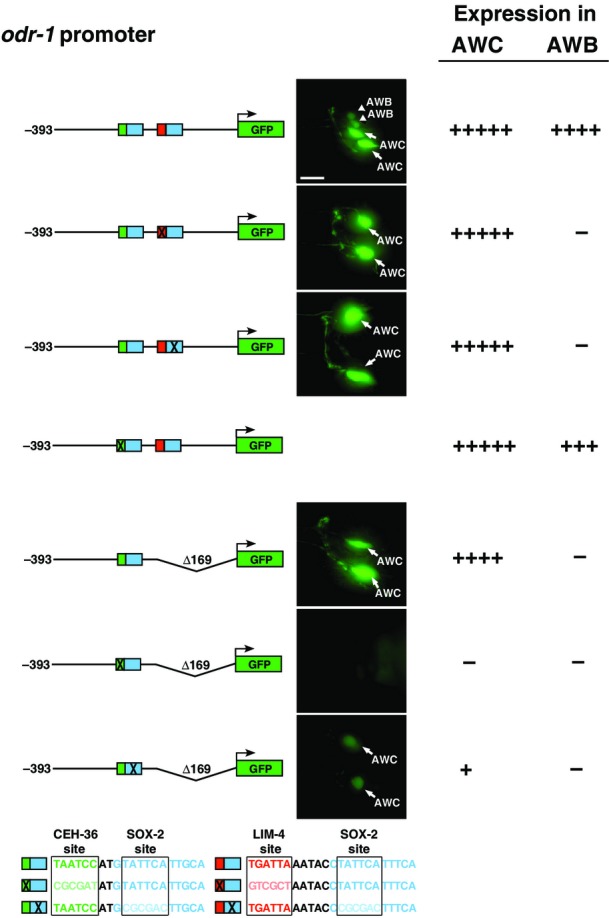
odr-1 promoter GFP reporter constructs and their expression levels in AWB and AWC cells
Increased number of (+) indicates higher intensity of GFP expression; (−) indicates lack of expression. Consensus binding sites of CEH-36, SOX-2, and LIM-4 are boxed. Green, CEH-36 site; blue, SOX-2 site; red, LIM-4 site. Lighter shades of green, blue, and red as well as X represent mutated sites. Scale bar, 10 μm.
To corroborate that these transcription factors directly bind to their respective predicted target sites, we performed electrophoretic mobility shift assays (EMSAs) using 6×His-tagged proteins and IRDye-labeled probes containing either the CEH-36/SOX-2 or LIM-4/SOX-2 adjacent sites (Appendix Fig S1). Our results show specific binding of SOX-2 to both the CEH-36/SOX-2 and LIM-4/SOX-2 probes, as wild-type unlabeled probes were more efficient at competing with labeled probes in forming the protein–DNA complex than unlabeled probes with mutated SOX-2 binding sites (Appendix Fig S1A and B). CEH-36 and LIM-4 also showed specific binding to the CEH-36/SOX-2 and LIM-4/SOX-2 probes, respectively, since wild-type unlabeled probes were better competitors than the unlabeled probes with a mutated CEH-36 or LIM-4 site (Appendix Fig S1C and D). Both SOX-2 and SOX-2G73E caused a similar degree of shift of the CEH-36/SOX-2 probe (Fig7E), indicating that the SOX-2G73E mutation does not affect DNA binding to the SOX-2 site in the CEH-36/SOX-2 element. However, SOX-2G73E showed a much reduced level, compared with wild-type SOX-2, in binding to the LIM-4/SOX-2 probe (Fig7C). To determine the ability of SOX-2 and SOX-2G73E to form a complex with LIM-4 or CEH-36 together with DNA, supershift assays were performed. SOX-2 was more efficient at forming a complex with the LIM-4/SOX-2 probe and LIM-4 protein (Fig7D, lane 3) than SOX-2G73E (Fig7D, lane 5). However, SOX-2 and SOX-2G73E had similar levels of ability in forming a complex with CEH-36 and the CEH-36/SOX-2 probe (Fig7F, lanes 3, 5). Together, these results further suggest a model in which SOX-2 functions cooperatively with both CEH-36 and LIM-4 in a DNA-dependent manner, while SOX-2G73E can still cooperate with CEH-36 through binding to the CEH-36/SOX-2 element but not with LIM-4 due to its reduced binding ability to the LIM-4/SOX-2 element (Fig8).
Lastly, we note that the sox-2(ky707) allele appears to selectively affect the function of sox-2 in AWB specification (via interaction with LIM-4) since sox-2(ky707) mutant animals do not display any of the additional pleiotropies associated with complete loss of sox-2 function which we recently described [larval lethality, specification of OLL and IL1 sensory neurons (Vidal et al, 2015)].
Maintained expression of sox-2,lim-4, and ceh-36 depends on auto- and cross-regulation
sox-2,lim-4, and ceh-36 are continuously expressed from embryonic to adult stages, which is consistent with the role of these transcription factors in the initiation of a terminal differentiation program and the maintenance of the differentiated state. It was previously shown that lim-4 autoregulates its expression in AWB and ceh-36 autoregulates in AWC (Appendix Fig S2) (Sagasti et al, 1999; Kim et al, 2010). To determine (i) whether sox-2 also autoregulates, and (ii) whether these transcription factors also cross-regulate, we examined the expression of each factor in sox-2(null),sox-2(ky707),ceh-36(lf), and lim-4(lf) mutants (Appendix Fig S2).
The expression of the sox-2ps::2xnlsGFP reporter gene was significantly reduced in AWC neurons in sox-2(ot640) null mutants as well as in AWB neurons in sox-2(ot640) and sox-2(ky707) mutants (Appendix Fig S2), suggesting that sox -2 regulates its own expression in both AWC and AWB. In addition, the expression of sox-2ps::2xnlsGFP was significantly reduced in AWC in ceh-36(ky640lf) mutants and in AWB in lim-4(ky403lf) mutants (Appendix Fig S2), suggesting that ceh-36 and lim-4 are required for sox-2 expression in AWC and AWB, respectively. Furthermore, the expression of a ceh-36p::GFP reporter gene in AWC and a lim-4p::GFP reporter gene in AWB was significantly reduced in sox-2(ot640) null mutants (Appendix Fig S2), suggesting that sox-2 regulates ceh-36 and lim-4 expression in AWC and AWB, respectively. Together, these results suggest that maintained expression of sox-2,lim-4, and ceh-36 is achieved through autoregulation and cross-regulation.
Gene dosage experiments suggest a competition model
The data presented so far demonstrate two main features of AWB and AWC differentiation (schematically summarized in Fig8): (i) SOX-2 and LIM-4 cooperate in AWB to induce AWB identity and to prevent the adoption of the alternative AWC identity by repressing the expression of the ceh-36 gene, an inducer of AWC identity, and (ii) CEH-36 cooperates with SOX-2 to induce AWC identity in AWC neurons of wild-type animals or in AWB in lim-4 or sox-2(ky707) mutants (in which the cooperation of SOX-2 with LIM-4 is affected). Since sox-2 is required for both the AWB and AWC differentiation programs, we considered a competition model. In this model, LIM-4 may repress the alternative AWC identity in AWB by preferentially cooperating with SOX-2, thereby limiting the access of CEH-36 to SOX-2. Since SOX-2 and CEH-36 are required for ceh-36 expression itself, the ultimate output of LIM-4's preferential cooperation with SOX-2 is a failure to express ceh-36 and hence AWC identity. Although we are unable to observe ceh-36 expression in AWB neurons in wild-type animals, it may be transiently expressed in embryonic AWB neurons and suppressed fairly quickly by lim-4.
We tested the competition model by probing specific predictions of the model. First, this model would predict that ectopic expression of lim-4 in the AWC neurons is sufficient to induce AWB identity in AWC and to repress ceh-36 expression and hence AWC identity. We found that ectopic lim-4 expression in AWC from the odr-3p::lim-4 transgene indeed repressed ceh-36 expression in AWC (Fig1B), resulting in loss of terminal AWC identity (Fig1B) (Sagasti et al, 1999). In addition, ectopic lim-4 expression in AWC induced the expression of AWB markers str-1 and lim-4 (Fig1B), as previously reported (Sagasti et al, 1999). Moreover, the ectopic induction of AWB identity by ectopic lim-4 expression in AWC genetically required the presence of wild-type sox-2 (Fig1B).
Vice versa, the competition model predicts that overexpression of ceh-36 in AWB from the odr-3p::ceh-36 transgene may be able to promote CEH-36/SOX-2 cooperation in AWB and thereby overcome the normally preferred LIM-4/SOX-2 cooperation, resulting in the induction of AWC identity in AWB and loss of AWB identity. We found this to be indeed the case: The AWC marker str-2 was induced and the AWB markers str-1 and lim-4 were reduced (Fig5B).
Lastly, the competition model predicts that SOX-2 is present in rate-limiting amounts in AWB, so that all available SOX-2 cooperates only with LIM-4. Raising the level of SOX-2 in AWB should make SOX-2 become available for cooperation with CEH-36. This should result in the induction of AWC identity in AWB, but in this case, normal AWB identity should be unaffected because LIM-4 can still cooperate with SOX-2 to also induce AWB identity. We indeed observed a mixed AWB/AWC identity of AWB neurons in animals that expressed sox-2 under control of the odr-3 promoter from the odr-3p::sox-2 transgene: The AWC marker str-2 was induced and the AWB markers str-1 and lim-4 were unaffected (Fig5B). This also suggests that overexpression of sox-2 may be able to bypass lim-4 repression of ceh-36 in AWB neurons.
Taken together, even though we cannot rule out alternative scenarios, our data suggest that SOX-2 can partner with both LIM-4 and CEH-36, but preferential partnering of LIM-4 with SOX-2 is able to overcome a CEH-36/SOX-2-driven “AWC ground state” that would normally be executed in AWC and AWB in the absence of lim-4.
Distinct differentiation mechanisms for distinct olfactory neuron types
One key aspect of the sox-2-dependent AWB/AWC differentiation event is the concept of a ceh-36/sox-2-dependent “AWC ground state” that is modified by lim-4 to drive AWB and inhibit AWC identity in a sox-2 dependent manner. Several previous studies have suggested that AWC is also a default identity state for several other amphid sensory neuron classes, as this ground state is revealed upon removal of the terminal selector gene of the respective amphid sensory identity. Specifically, loss of AWA identity in odr-7 or lin-11 mutants, loss of ASG identity in unc-130; odr-7 double mutants, or loss of AFD identity in ttx-1 mutants has been proposed to reveal an AWC ground state (Sagasti et al, 1999; Sarafi-Reinach & Sengupta, 2000; Sarafi-Reinach et al, 2001; Satterlee et al, 2001; Lanjuin et al, 2003). However, these conclusions were mainly if not exclusively based on the ectopic expression of the GPCR str-2, an AWCON marker. str-2 has been found to be exquisitely sensitive to various sensory signals and sensory activity (Peckol et al, 2001; Nolan et al, 2002), and the change in str-2 expression may therefore not necessarily reflect a cell identity switch. We revisited this issue by examining whether another AWCON marker srt-28 (GPCR gene), the AWC terminal identity marker odr-1 (guanylyl cyclase gene), and the AWC terminal selector ceh-36 (Otx-homeobox gene) are indeed derepressed if the identity of other amphid sensory neurons is lost, as would be expected from the AWC default model. We found that only the srt-28 marker, like str-2, was ectopically expressed in AWA in odr-7 mutants, but no evidence of identity switches from other amphid sensory neurons to AWC in odr-7, lin-11, or ttx-1 mutants when odr-1 and ceh-36 markers were examined (Appendix Fig S3). These results collectively suggest that AWC is not the default state of AWA, ASG, or AFD neurons. Taken together with previous studies, our findings indicate that olfactory sensory neurons are specified in fundamentally distinct manners. AWB and AWC share a common ground state, even though these neurons are lineally unrelated, but other olfactory (AWA), gustatory (ASG), or thermosensory (AFD) neurons do not develop from this common “ground state,”
Discussion
We have identified a novel role of sox-2 in postmitotic specification and differentiation of two distinct, but related types of sensory neurons, the olfactory AWB and AWC neuron types, in C. elegans. Previous work has demonstrated a role of sox-2 in controlling the terminal differentiation of postmitotic neuron types (Vidal et al, 2015), yet the mechanistic basis for how sox-2 fulfills such function has been unclear. We define here a competition mechanism by which two distinct homeodomain factors can compete for cooperation with sox-2 to drive neuron-type-specific gene expression programs.
Several studies have shown that cooperative interactions of vertebrate Sox2 with various partner factors determine target gene specificity, thereby providing a code of cell specification (Kamachi & Kondoh, 2013). For example, Sox2 interaction with Brn2 defines a neural progenitor state (Lodato et al, 2013), the Sox2-Pax6 partnership specifies lens development (Kamachi et al, 2001), Sox2 and Gli cooperate in Shh-directed neural tube patterning (Peterson et al, 2012), and Sox2 functions with Tcf/Lef to repress cyclin D1 (Hagey & Muhr, 2014). Our results suggest a model in which SOX-2 cooperates with either LIM-4 or CEH-36 to distinguish between the AWB versus the AWC olfactory neuron identity (Fig8). In AWC neurons, SOX-2 cooperates with CEH-36 as a terminal selector to activate transcription of AWC-specific genes through binding to the CEH-36/SOX-2 adjacent sites (Fig8A). In AWB neurons, SOX-2 pairs with LIM-4 to act as a terminal selector for AWB identity by binding to the LIM-4/SOX-2 adjacent sites (Fig8A). The pairing of LIM-4 and CEH-36 with SOX-2 generates unique combinatorial transcription factor codes that define the specificity of action of LIM-4 and CEH-36, each of which is expressed in additional, different neuron types that do not express SOX-2.
In addition, lim-4 represses ceh-36 expression and therefore prevents the execution of the alternative AWC identity in AWB (Fig8A). This repression may be direct or it may be the result of a competition of LIM-4 with CEH-36 for cooperation with SOX-2, with LIM-4 being the preferred SOX-2 partner, thereby preventing SOX-2/CEH-36-mediated AWC induction. Gene dosage experiments that we performed favor the competition model.
The sox-2(ky707) mutation causes a glycine to glutamic acid change in the highly conserved HMG-box domain. The HMG-box domain of Sox proteins has multiple important roles in the regulation of target gene expression through binding to DNA and/or interacting with different partner proteins (Lefebvre et al, 2007). The AWB-to-AWC transformation phenotype resulting from the sox-2(ky707) mutation suggests that the mutation may change the binding specificity of SOX-2 with the target genes and/or its interacting proteins in AWB neurons. Our electrophoretic mobility shift assay results suggest the mutant SOX-2G73E can still cooperate with CEH-36 through binding to the CEH-36/SOX-2 element, while SOX-2G73E is no longer able to cooperate with LIM-4 due to its reduced binding ability to the LIM-4/SOX-2 element. This leads to the derepression of the SOX-2/CEH-36 partnership in AWB cells, causing them to transform into AWC neurons (Fig8B). When SOX-2 is abolished, as in sox-2(ot640) null mutants, both AWB and AWC identities are lost (Fig8C). Taken together, CEH-36 and SOX-2 define an “AWC ground state” on which the AWB identity is superimposed by LIM-4 cooperating with SOX-2, which induces AWB identity and represses AWC identity. “AWC identity” is the ground state only for the AWB and AWC neurons, and it is not a general ground state for other olfactory and sensory neurons as previously proposed (Sagasti et al, 1999; Sarafi-Reinach et al, 2001; Lanjuin et al, 2003).
The competition mechanism by which AWB and AWC neurons are specified bears striking similarities to the specification of the ALM and BDU neurons that we recently described (Gordon & Hobert, 2015). In both cases, two neurons share a common terminal selector (SOX-2 in AWB and AWC; UNC-86 in ALM and BDU) that pairs up with a distinct cofactor to distinguish between identities of different neuron types (SOX-2/LIM-4 pair in AWB and SOX-2/CEH-36 pair in AWC; UNC-86/MEC-3 pair in ALM and UNC-86/PAG-3 pair in BDU). Another similarity observed is that one of the cofactors (LIM-4 in AWB and MEC-3 in ALM) outcompetes the other potential cofactor (CEH-36 in AWB and PAG-3 in ALM) for cooperation with the terminal selector. Hence, the loss of either cofactor (LIM-4 in AWB and MEC-3 in ALM) results in what essentially amounts to a homeotic transformation in cellular identity (AWB to AWC as described here; ALM to BDU as described in Gordon and Hobert (2015)). Having observed such a mechanism in two completely different cellular contexts suggests that it may be a common principle that distinct neuron types may share a common terminal selector, which pair up with distinct cofactors in a competitive manner. In an evolutionary context, this principle may mean that neuron identities may diversify from an ancestral ground state through the recruitment of a terminal selector into a new cellular context where it can cooperate with a resident terminal selector to drive a novel cellular identity.
Since a number of SOX and homeobox genes are highly expressed in adult human olfactory bulb (Marei et al, 2012) and since the binding motifs of homeodomain proteins and SOX family proteins are overrepresented in the promoters of mouse olfactory receptor genes (Plessy et al, 2012), we propose that the role of Sox2 in olfactory neuron specification may be conserved in vertebrates and that Sox2 may operate with as yet to be identified homeodomain proteins to fulfill this function in vertebrates.
Materials and Methods
Strains and transgenes
Wild-type is strain N2, C. elegans variety Bristol. Strains were maintained by standard methods (Brenner, 1974). Animal care and use were carried out in accordance with the Institutional Biosafety Committee Policies. A list of strains and transgenes can be found in the Appendix Supplementary Methods. Power analysis was used to determine the sample size that is adequate to detect a significant effect size.
Forward genetic screen
Forward genetic screen was performed as previously described (Brenner, 1974). P0 animals were mutagenized with EMS. Five F1 progenies were then transferred onto single plates, and F2 worms were screened for mutants using the Zeiss fluorescence compound microscope. The ky707 mutant was identified from a screen of 2,400 genomes.
Genetic mapping and whole-genome sequencing
Three-factor mapping experiments were performed to map ky707 to chromosome X between unc-6(n102) (X:-2.01) and dpy-7(e88) (X:-1.65). Whole-genome sequencing of the ky707 mutant was performed on an Illumina GA2x sequencing platform, using 100-nucleotide reads. Sequencing results were analyzed with the MAQGene software as previously described (Bigelow et al, 2009).
Plasmid construction
A list of plasmid DNA can be found in the Appendix Supplementary Methods.
Behavioral assays
Chemotaxis assays were performed and chemotaxis indices were computed as in previous studies (Bargmann et al, 1993; Troemel et al, 1997). 2-butanone (Sigma 360473) and 2,3-pentanedione (Sigma 241962) were diluted 1:1,000 and 1:10,000, respectively, in ethanol. About 50-150 animals were assayed for each strain and each odor in each individual assay. All assays were performed four independent times.
Mammalian cell transfection and luciferase assays
Mammalian expression constructs were transfected into COS-7 cells using the Lipofectamine 2000 reagent (Invitrogen) in a 24-well format according to the manufacture's protocol. A control vector (gift of J. Wells, Cincinnati Children's) expressing Renilla Luciferase from the CMV promoter was used for normalizing transfection efficiency. The total amount of DNA was kept constant with addition of the pcDNA6 V5 6×His A empty vector. Cell lysates were prepared 48 h after transfection, and luciferase activity was measured using the Firefly & Renilla Luciferase Assay Kit (Biotium) and GloMax-96 Microplate Luminometer (Promega). All transfection assays were performed in triplicate and repeated three times.
6×His-tagged protein expression and purification
To make 6×His-tagged proteins, sox-2,sox-2G73E, lim-4, and ceh-36 were cloned into pRSETA (Invitrogen) and transformed into BL21-CodonPlus(DE3)-RIL bacterial cells (Stratagene). IPTG (0.1 or 1 mM) was added in transformed cell culture (OD600 = ∼0.7) to induce 6×His-tagged protein expression for 1–4 h at 37°C. Cells were then harvested and lysed with B-PER in phosphate buffer (Thermo Scientific Pierce). 6×His-tagged proteins were purified using a HisPur Cobalt Purification Kit (Thermo Scientific Pierce). SDS–PAGE and Coomassie staining were used to determine the purity and concentration of purified 6×His-tagged proteins.
Electrophoretic mobility shift assay (EMSA)
To make IRDye-labeled probes, long oligonucleotides (51–52-mers, IDT) and complementary short oligonucleotides (13–14-mers, IDT) with 5′ IR-700 dye modification were annealed, and 5′ overhangs were filled in with Klenow DNA polymerase (NEB). Long oligonucleotides of complementary sequences were annealed to make unlabeled probes for competition analysis with IRDye-labeled probes. Purified 6×His-tagged proteins (80–200 ng), unlabeled probes (10–125 nM, only added for competition experiments in Appendix Fig S1), and IRDye-labeled probes (5 nM) were incubated at room temperature in 1× binding buffer (10 mM Tris–HCl pH 7.5, 10 mM NaCl, 40 mM KCl, 1 mM MgCl2, 1 mM EDTA pH 8.0, 5% glycerol, and 50 μg/ml BSA) for 15 min and resolved in a pre-run 5% polyacrylamide gel containing 2.5% glycerol in 0.5× TBE buffer. The small gel (8.3 cm width × 7.3 cm length × 0.75 mm thickness, Bio-Rad Mini-PROTEAN; Fig7C and E; Appendix Fig S1) was run at 80 V for up to 75 min, and the large gel (16 cm width × 14 cm length × 1.5 mm thickness, Thermo Scientific Owl P9DS dual gel vertical electrophoresis; Fig7D and F) was run for 3 h. The gel was imaged using a LICOR Odyssey CLx Infrared Imaging System.
Acknowledgments
This work was supported by a Choose Ohio First Scholarship (A.A.), a National Institutes of Health (NIH) Organogenesis Training Grant (Y.-W.H.), the NSF (IOS-1455758 to C.C.), the March of Dimes Foundation (C.C.), Whitehall Foundation Research Awards (C.C. and C.-F.C), Alfred P. Sloan Research Fellowship (C.-F.C.), the Howard Hughes Medical Institute (O.H.), and the NIH (5R01GM111320-02 to C.C., R01NS039996-05 and R01NS050266-03 to O.H., and 5R01GM098026-05 to C.-F.C.). The ky707 allele was isolated by C.-F.C. as a postdoctoral fellow in Cori Bargmann's laboratory. We thank Cori Bargmann and Piali Sengupta for neuronal marker strains, Alex Boyanov for whole-genome sequencing of the ky707 mutant, Maria Doitsidou for assistance in MAQGene analysis of whole-genome sequencing results, Kyle McCracken and Jim Wells for advice on cell transfection and luciferase assays, Brian Gebelein for advice on EMSAs, Yutaka Yoshida for mouse cDNA, Andy Fire for C. elegans vectors, and C. elegans Genetic Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for strains.
Author contributions
AA, Y-WH, BV, CC, OH, and C-FC conceived or designed the experiments. AA, Y-WH, BV, and C-FC performed the experiments. AA, Y-WH, BV, CC, OH, and C-FC analyzed the data. AA, Y-WH, OH, and C-FC wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Appendix
Expanded View Figures PDF
Review Process File
References
- Ambrosetti DC, Scholer HR, Dailey L, Basilico C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem. 2000;275:23387–23397. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. (2006) Chemosensation in C. elegans. WormBook, , ed. The C. elegans Research Community, WormBook, doi: 10.1895/wormbook.1.123.1 [DOI] [PMC free article] [PubMed]
- Bauer Huang SL, Saheki Y, VanHoven MK, Torayama I, Ishihara T, Katsura I, van der Linden A, Sengupta P, Bargmann CI. Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural Dev. 2007;2:24. doi: 10.1186/1749-8104-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow H, Doitsidou M, Sarin S, Hobert O. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat Methods. 2009;6:549. doi: 10.1038/nmeth.f.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Hsieh YW, Lesch BJ, Bargmann CI, Chuang CF. Microtubule-based localization of a synaptic calcium-signaling complex is required for left-right neuronal asymmetry in C. elegans. Development. 2011;138:3509–3518. doi: 10.1242/dev.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Vanhoven MK, Fetter RD, Verselis VK, Bargmann CI. An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell. 2007;129:787–799. doi: 10.1016/j.cell.2007.02.052. [DOI] [PubMed] [Google Scholar]
- Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, Szumska D, Kockx C, van Ijcken W, Dekkers DH, Demmers J, Rijkers EJ, Bhattacharya S, Philipsen S, Pevny LH, Grosveld FG, Rottier RJ, Lenhard B, Poot RA. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G, Lin B. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genom. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein JL. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PM, Hobert O. A Competition Mechanism for a Homeotic Neuron Identity Transformation in C. elegans. Dev Cell. 2015;34:206–219. doi: 10.1016/j.devcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth SI, Wegner M. Having it both ways: Sox protein function between conservation and innovation. Cell Mol Life Sci. 2008;65:3000–3018. doi: 10.1007/s00018-008-8138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagey DW, Muhr J. Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 2014;9:1908–1920. doi: 10.1016/j.celrep.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Hobert O. (2010) Neurogenesis in the nematode Caenorhabditis elegans. WormBook, ed. The C. elegans Research Community, WormBook, doi: 10.1895/wormbook.1.12.2
- Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- Hsieh YW, Alqadah A, Chuang CF. Asymmetric neural development in the Caenorhabditis elegans olfactory system. Genesis. 2014;52:544–554. doi: 10.1002/dvg.22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- Kazemian M, Pham H, Wolfe SA, Brodsky MH, Sinha S. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013;41:8237–8252. doi: 10.1093/nar/gkt598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Kim R, Sengupta P. The HMX/NKX homeodomain protein MLS-2 specifies the identity of the AWC sensory neuron type via regulation of the ceh-36 Otx gene in C. elegans. Development. 2010;137:963–974. doi: 10.1242/dev.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lanjuin A, Sengupta P. Specification of chemosensory neuron subtype identities in Caenorhabditis elegans. Curr Opin Neurobiol. 2004;14:22–30. doi: 10.1016/j.conb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Gehrke AR, Bulyk ML, Bargmann CI. Transcriptional regulation and stabilization of left-right neuronal identity in C. elegans. Genes Dev. 2009;23:345–358. doi: 10.1101/gad.1763509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Bargmann CI. The homeodomain protein hmbx-1 maintains asymmetric gene expression in adult C. elegans olfactory neurons. Genes Dev. 2010;24:1802–1815. doi: 10.1101/gad.1932610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Etoile ND, Bargmann CI. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron. 2000;25:575–586. doi: 10.1016/s0896-6273(00)81061-2. [DOI] [PubMed] [Google Scholar]
- Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet. 2013;9:e1003288. doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes R, Verhey van Wijk N, Neves G, Pachnis V. Transcription factor LIM homeobox 7 (Lhx7) maintains subtype identity of cholinergic interneurons in the mammalian striatum. Proc Natl Acad Sci USA. 2012;109:3119–3124. doi: 10.1073/pnas.1109251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marei HE, Ahmed AE, Michetti F, Pescatori M, Pallini R, Casalbore P, Cenciarelli C, Elhadidy M. Gene expression profile of adult human olfactory bulb and embryonic neural stem cell suggests distinct signaling pathways and epigenetic control. PLoS One. 2012;7:e33542. doi: 10.1371/journal.pone.0033542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KM, Sarafi-Reinach TR, Horne JG, Saffer AM, Sengupta P. The DAF-7 TGF-beta signaling pathway regulates chemosensory receptor gene expression in C. elegans. Genes Dev. 2002;16:3061–3073. doi: 10.1101/gad.1027702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H. Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development. 2005;132:1663–1674. doi: 10.1242/dev.01707. [DOI] [PubMed] [Google Scholar]
- Peckol EL, Troemel ER, Bargmann CI. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2001;98:11032–11038. doi: 10.1073/pnas.191352498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, McFarlane M, Baizabal JM, Junker JP, van Oudenaarden A, Mikkelsen T, Bernstein BE, Bailey TL, Bulyk ML, Wong WH, McMahon AP. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, Vassalli A, Lazarevic D, Severin J, Vlachouli C, Simone R, Faulkner GJ, Kawai J, Daub CO, Zucchelli S, Hayashizaki Y, Mombaerts P, Lenhard B, Gustincich S, Carninci P. Promoter architecture of mouse olfactory receptor genes. Genome Res. 2012;22:486–497. doi: 10.1101/gr.126201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rister J, Desplan C, Vasiliauskas D. Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development. 2013;140:493–503. doi: 10.1242/dev.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 1999;13:1794–1806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasti A, Hisamoto N, Hyodo J, Tanaka-Hino M, Matsumoto K, Bargmann CI. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 2001;105:221–232. doi: 10.1016/s0092-8674(01)00313-0. [DOI] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach TR, Sengupta P. The forkhead domain gene unc-130 generates chemosensory neuron diversity in C. elegans. Genes Dev. 2000;14:2472–2485. doi: 10.1101/gad.832300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach TR, Melkman T, Hobert O, Sengupta P. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development. 2001;128:3269–3281. doi: 10.1242/dev.128.17.3269. [DOI] [PubMed] [Google Scholar]
- Satterlee JS, Sasakura H, Kuhara A, Berkeley M, Mori I, Sengupta P. Specification of thermosensory neuron fate in C. elegans requires ttx-1, a homolog of otd/Otx. Neuron. 2001;31:943–956. doi: 10.1016/s0896-6273(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Schumacher JA, Hsieh YW, Chen S, Pirri JK, Alkema MJ, Li WH, Chang C, Chuang CF. Intercellular calcium signaling in a gap junction-coupled cell network establishes asymmetric neuronal fates in C. elegans. Development. 2012;139:4191–4201. doi: 10.1242/dev.083428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Colbert HA, Bargmann CI. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell. 1994;79:971–980. doi: 10.1016/0092-8674(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, Hobert O. Modular Control of Glutamatergic Neuronal Identity in C. elegans by Distinct Homeodomain Proteins. Cell. 2013;155:659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Hino M, Sagasti A, Hisamoto N, Kawasaki M, Nakano S, Ninomiya-Tsuji J, Bargmann CI, Matsumoto K. SEK-1 MAPKK mediates Ca2 + signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Hsieh YW, Gamse JT, Chuang CF. Making a difference together: reciprocal interactions in C. elegans and zebrafish asymmetric neural development. Development. 2010;137:681–691. doi: 10.1242/dev.038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Troemel ER. Chemosensory signaling in C. elegans. BioEssays. 1999;21:1011–1020. doi: 10.1002/(SICI)1521-1878(199912)22:1<1011::AID-BIES5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- Vanhoven MK, Bauer Huang SL, Albin SD, Bargmann CI. The claudin superfamily protein nsy-4 biases lateral signaling to generate left-right asymmetry in C. elegans olfactory neurons. Neuron. 2006;51:291–302. doi: 10.1016/j.neuron.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Vidal B, Santella A, Serrano-Saiz E, Bao Z, Chuang CF, Hobert O. C. elegans SoxB genes are dispensable for embryonic neurogenesis but required for terminal differentiation of specific neuron types. Development. 2015;142:2464–2477. doi: 10.1242/dev.125740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Bargmann CI. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File