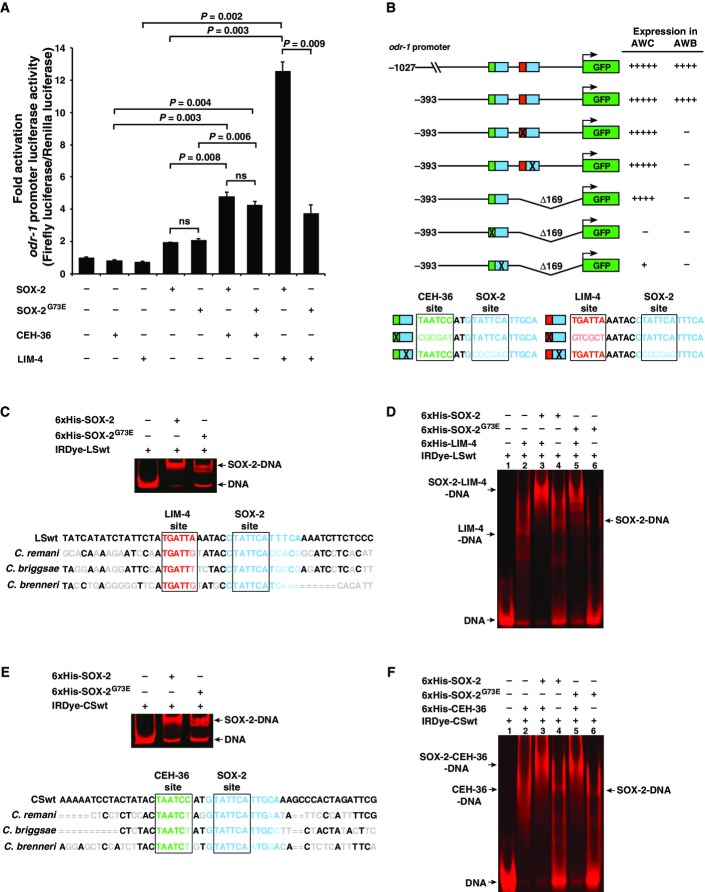
Figure 7.
- A Quantification of fold activation of odr-1 promoter measured using dual-spectral luciferase assays in COS-7 cells. The 1,027-bp odr-1 promoter, expressed in both AWB and AWC neurons, has basal activity in the presence of the single transcription factors CEH-36, LIM-4, SOX-2, or SOX-2G73E. The activity is boosted significantly in the presence of both CEH-36/SOX-2 and LIM-4/SOX-2 combinations. The sox-2(ky707) mutation disrupts the cooperation between LIM-4 and SOX-2G73E, but does not affect CEH-36/SOX-2G73E cooperation. All transfection assays were performed in triplicate and repeated three times; three repeats showed similar trends of results. Data of one set of triplicate are presented. Unpaired Student's t-test was used to determine P-value; ns, not significant.
- B odr-1 promoter GFP reporter constructs and their expression levels in AWB and AWC cells. Increased number of (+) indicates higher intensity of GFP expression; (−) indicates lack of expression. Consensus binding sites of CEH-36, SOX-2, and LIM-4 are boxed. Green, CEH-36 site; blue, SOX-2 site; red, LIM-4 site. Lighter shades of green, blue, and red as well as X represent mutated sites.
- C–F EMSA assays of 6×His-tagged proteins with an IRDye-labeled DNA probe containing LIM-4/SOX-2 adjacent binding sites (CSwt) (C, D) or CEH-36/SOX-2 adjacent binding sites (LSwt) (E, F). Nucleotides in gray, light green, light blue, and light red are sequences of C. remani,C. briggsae, and C. brenneri that differ from C. elegans sequence. = indicates no alignment available between different species. Sequence alignment between species was performed on http://genome.ucsc.edu. Gel images in (D) and (F) were spliced between lanes 2–3 and 4–5 to exclude irrelevant lanes.