Figure 2. Smc5/6 regulates Mph1 and recombination independently of Rtt107.
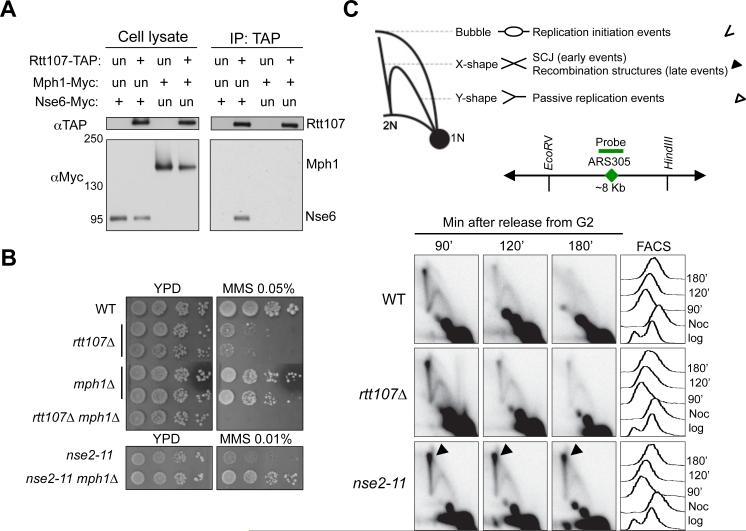
(A) Rtt107 interacts with the Nse6 subunit of the Smc5/6 complex, but not the DNA helicase Mph1. Experiments were carried out and results are presented as in Figure 1A.
(B) mph1Δ suppresses the MMS sensitivity of the nse2 mutant but not that of rtt107Δ. Experiments were done as in Figure 1D, except that cells were spotted on normal media (YPD) or YPD containing the indicated amount of MMS.
(C) Unlike nse2 mutants, rtt107Δ cells do not accumulate recombination intermediates. Top, schematic that depicts DNA structures visualized by 2D gel. Symbols for each structure are indicated at the right. Bubble DNA represents replication initiation structures; Y-shaped structures represent passive replication intermediates; X-shaped DNA are either sister chromatin junctions (SCJs) that form at an early time point, or other recombination structures that appear later in S phase. Middle, schematic for ARS305, probe position, and the restriction enzyme sites used for 2D gel analysis. Bottom, 2D gel and FACS results. Cells were arrested in nocodazole and released into medium containing 0.03% MMS, and samples were examined at the indicated time points.
See also Figure S2.