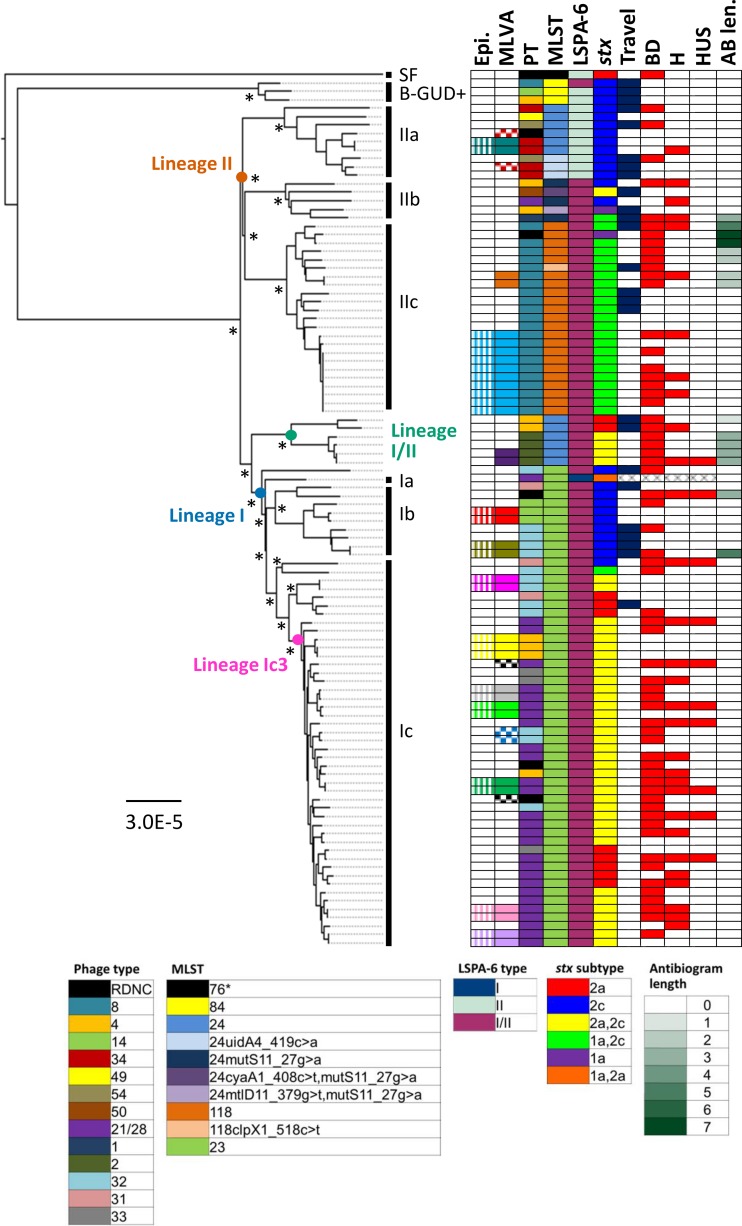
FIG 1.
Maximum-likelihood core genome phylogeny for 103 E. coli O157 isolates from Lothian, Scotland. Recombinant sequences XH18570E and XH22083W were excluded. The tree was constructed with RAxML by using a general time-reversible model of nucleotide substitution and gamma distributed rate heterogeneity across sites. Branch lengths are in numbers of substitutions per site. Isolates known to be epidemiologically related (Epi) are in the same color. MLVA types shared by more than one isolate are indicated by filled boxes of the same color; SLV MLVA types are indicated by checkered boxes of the same color. PTs, multilocus STs, LSPA-6 types, stx subtypes, and the number of antibiotics (out of 15 tested) to which an isolate was resistant (AB len.) are shown. RDNC indicates that the PT reaction did not conform to a recognized pattern. Isolates associated with recent travel are in blue (Travel). All Lothian isolates were associated with diarrhea; isolates associated with bloody diarrhea (BD), hospitalization (H), and/or HUS are in red. Sublineages, defined as described in the text, are indicated by vertical black bars. One thousand bootstrap replicates were conducted, and bootstrap values of >98%, for nodes corresponding to the major lineages and sublineages described in the text, are indicated by asterisks.