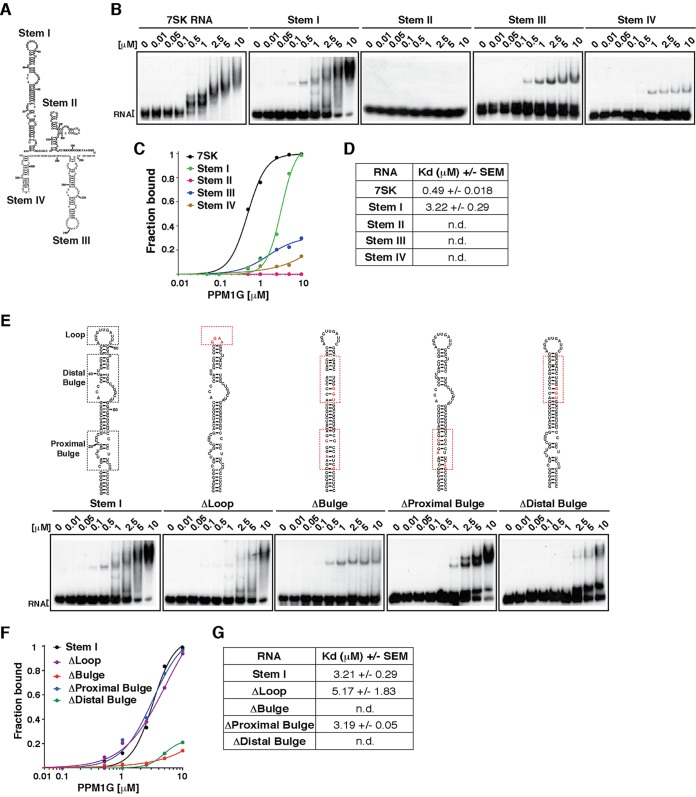
FIG 2.
PPM1G binds the 7SK RNA stem I distal bulge. (A) Primary sequence and predicted secondary structure of human 7SK RNA with the corresponding stem-loops (Stem). (B) Gel shift assays with full-length PPM1G and 7SK RNA or each of the individual stems. (C) Binding curves from the gel shifts shown in panel B. (D) Calculations of the Kdapp values from data shown in panel C. Kdapp values represent the averages of data from three independent experiments and the standard errors of the means (n = 3). (E) Gel shift assays with PPM1G and WT 7SK stem I or mutants (ΔLoop, ΔBulge, ΔProximal Bulge, and ΔDistal Bulge). The schemes above the gel shifts show the positions of the different RNA elements and the corresponding mutations. (F) Binding curves from the gel shifts shown in panel E. (G) Calculations of the Kdapp from data shown in panel F (means ± standard errors of the means; n = 3). n.d., not determined, because the data set does not enable accurate calculation of the Kdapp values.