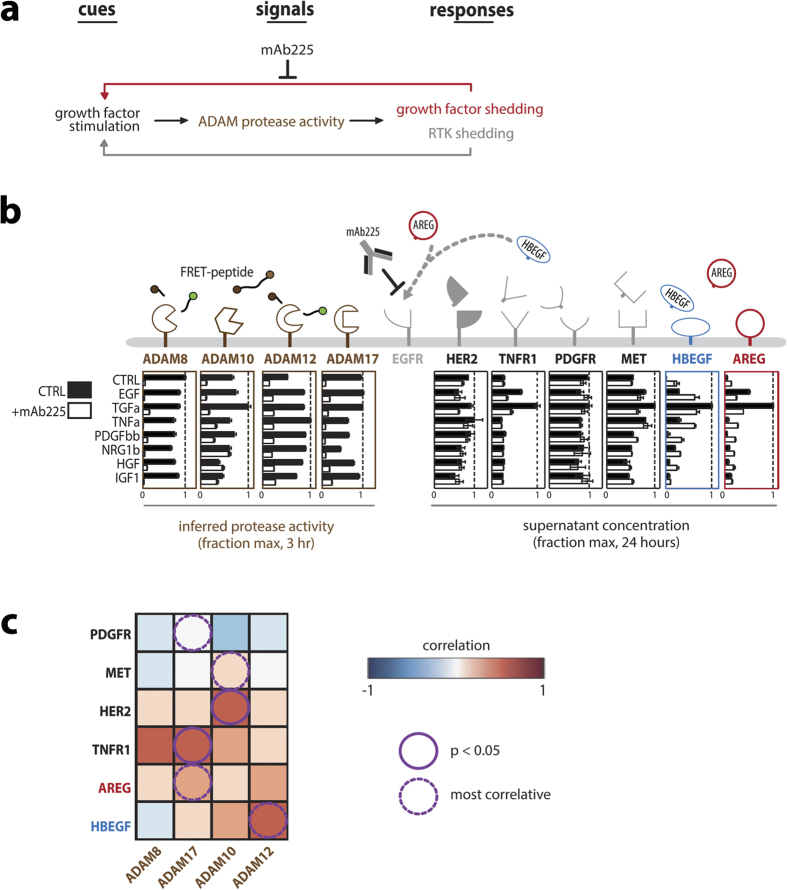
Figure 3. Cue-signal-response data set relates protease activities to ectodomain shedding in endometriosis cell culture.
(a) Overview schematic of cue-signal-response modeling approach to infer biochemical relationships between exogenously applied signaling cues, measured ADAM proteolytic activity “signals,” and corresponding ADAM-substrate shedding “responses.” (b) Serum-starved 12Z cells were treated with growth factor/cytokine “cues” for 3 h and simultaneously monitored in real-time for live-cell protease activity “signals” using FRET-based polypeptide probes. Left: Specific ADAM activities were then computationally inferred using the PrAMA algorithm (n = 4 ± SEM). Right: 24 h later, supernatants were analyzed using ELISA for endogenous ADAM-substrate accumulation “responses” (n = 3 reps ± SEM). (c) Pairwise Pearson correlations between protease activities ((b), left) and ADAM-substrate shedding ((b), right) were calculated for measurements as they varied across the panel of growth-factor/cytokine treatment conditions; results describe correlational relationships between patterns of inferred ADAM catalytic activity and downstream substrate proteolysis. EGF-ligand correlations were performed only for the n = 8 growth-factor treatment conditions in the presence of mAb225 to block ligand endocytosis, while receptor correlations were calculated with and without mAb225 (n = 16 conditions from n ≥ 3 reps; p-value from two-tailed t-tests).