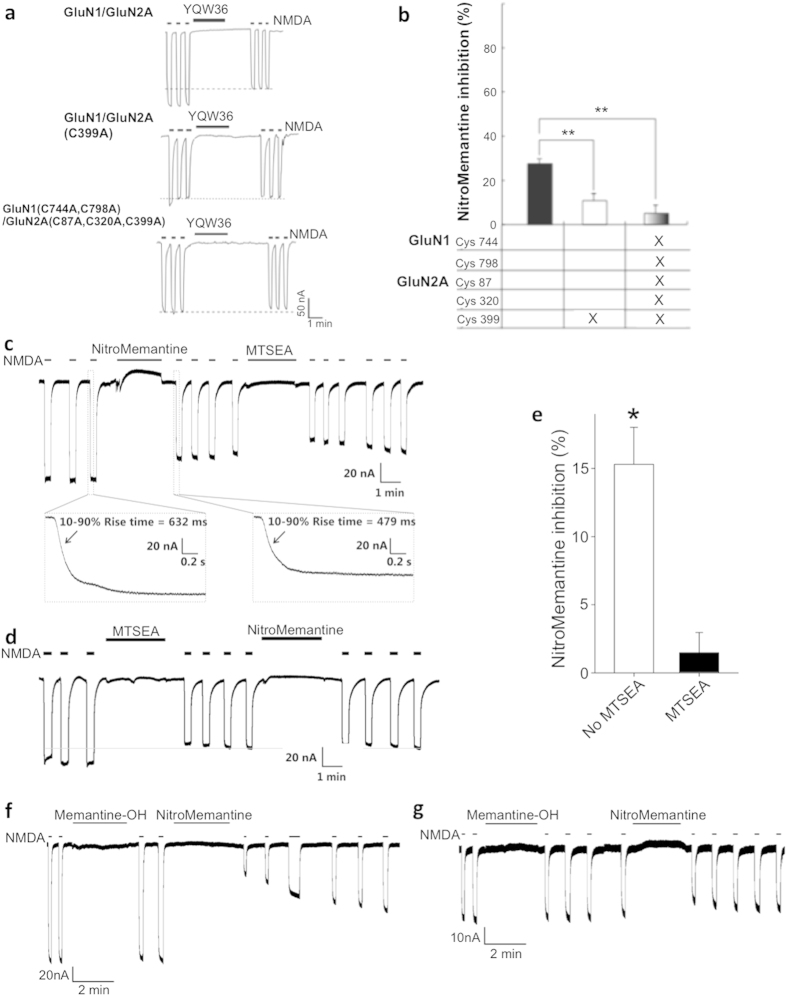
Figure 2. Redox effects of NitroMemantine YQW-036 mediated by S-nitrosylation.
(a) Representative recordings under two-electrode voltage clamp in oocytes expressing wild-type (WT) or non-nitrosylatable cysteine mutant NMDAR subunits. (b) Quantification of contribution of cysteine residues in various NMDAR subunits to the inhibitory effect of S-nitrosylation (data are mean + s.e.m. for n ≥ 5 recordings per data point, **P < 0.01). (c) In oocyte recordings of NMDA-evoked currents under voltage clamp, inhibition by NitroMemantine (10 μM) largely occluded any additional effect of subsequent application of the sulfhydryl-reactive reagent MTSEA (0.5 mM), as expected if the same cysteine residues were involved. Inset: expanded time scale to show rise time of NMDA-evoked current before and after NitroMemantine addition. (d) Inhibition of NMDA-evoked currents by MTSEA occludes the redox-mediated inhibitory effect of subsequent addition of NitroMemantine. (e) Quantification of effects shown in c and d. Data are mean + s.e.m. for n ≥ 6 oocytes recordings per data point (*P < 0.005). (f) NitroMemantine inhibits NMDAR activity by S-nitrosylation. Addition of 100 μM NitroMemantine YQW-036, but not its metabolite memantine-OH, inhibited NMDA-evoked current in oocytes expressing GluN1/GluN2A NMDARs recorded under voltage clamp conditions used to observe S-nitrosylation (Vh = −60 mV; data representative of n ≥ 4 oocytes recordings). (g) Mutation of the memantine binding site in the GluN1 subunit of the NMDAR prevents the redox effect of NitroMemantine. Disruption of the memantine binding site by mutation GluN1(N616R) abrogated the redox-mediated activity of NitroMemantine (100 μM). Data representative of n ≥ 3 oocytes recordings.