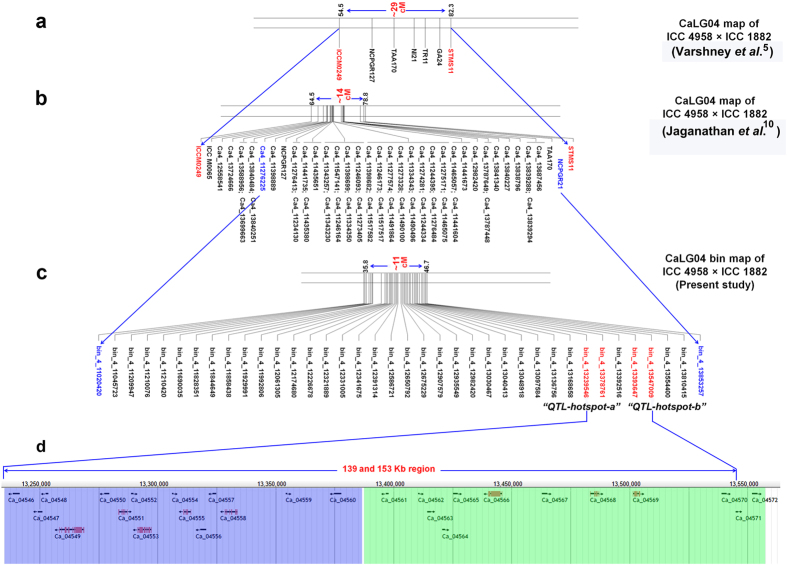
Figure 5. Refinement of “QTL-hotspot” region into “QTL-hotspot_a” and “QTL-hotspot_b” and identification of candidate genes.
(a) The “QTL-hotspot” region, reported by Varshney and colleagues5, spanning 29 cM (corresponds 7.74 Mb on physical map) harbouring QTLs for several drought tolerance related traits on CaLG04; (b) Refined “QTL-hotspot” region (~14 cM corresponding to ca. 3 Mb on physical map) reported by Jaganathan et al.10 consisted of 49 SNPs and six SSRs; (c) Refined “QTL-hotspot” region on CaLG04 with newly integrated markers (recombination bins) from the current study. The markers, viz. bin_4_13853257 and bin_4_11020420 correspond to the refined 3Mb “QTL-hotspot” region reported earlier10. Integration of 1,421 SNPs to the “QTL-hotspot” region resulted in the identification of 38 recombination breakpoints and thereby split the “QTL-hotspot” region into “QTL-hotspot_a” (139.22 Kb; 0.23 cM) and “QTL-hotspot_b” (153.36 Kb; 0.22 cM). The “QTL-hotspot_a” was flanked by bin_4_13239546 and bin_4_13378761 while “QTL-hotspot_b” was flanked by bin_4_13393647 and bin_4_13547009. These four flanking markers were shown in the red colour font; (d) A ~300 Kb (13,239,546-13,547,009) snapshot of chickpea genome from JBrowse showing twenty six candidate genes identified in the “QTL-hotspot_a” and “QTL-hotspot_b” regions. A total of 15 genes (highlighted in blue colour area) were identified from “QTL-hotspot_a” while 11 candidate genes (highlighted in green colour area) were identified from “QTL-hotspot_b” region.