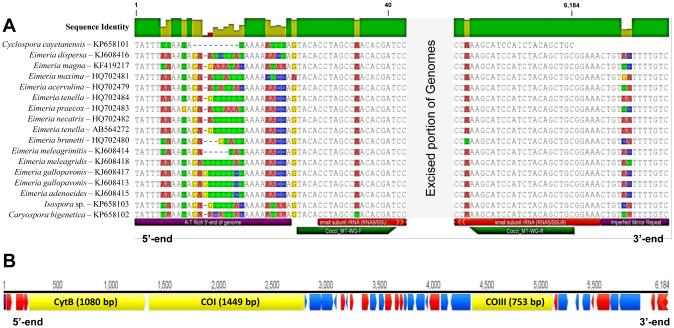
Fig. 1.
The mitochondrial (mt) genome of Cyclospora cayetanensis compared with the mt genomes of closely related eimeriid coccidia. (A) Aligned 5’- and 3’-termini of the Cyclospora cayetanensis mt genome aligned with the homologous regions in the mt genomes of other eimeriid coccidia; all comparative genomes have been linearized so that the first nucleotide of each mt genome matches the first base (‘T’) in the linear C. cayetanensis mt genome. A highly conserved region at the 5’-end with a consensus sequence of “5’-TATTTWWAADA…” was found in all genomes included in the alignment; all eimeriid genomes illustrated have the potential to form hairpins at the 5’-“end” of their circularly-mapped genomes. At the 3’-end of the eimeriid mt genome units in all coccidia other than C. cayetanensis (data not available) there was an imperfect mirror repeat sequence (i.e. …CTGTNHTTTTGTC -5’, conserved mirrored bases underlined in the preceding consensus sequence). The role, if any, of this terminal mirror repeat motif in the replication of eimeriid mt genomes is not known. The annotations listed below the Caryospora bigenetica mt genome sequence (terminal features, Cocci_MT-WG-F and R primers, and RNA9 and RNA5 ssrDNA fragments) apply to all sequences in the alignment. (B) Map of the C. cayetanensis mt genome illustrating the locations, directions and lengths of the three protein-coding genes (cytochrome b (CytB), cytochrome c oxidase subunit I (Cox1) and cytochrome c oxidase subunit III (Cox3)). The A-T rich 5’ end of the genome found in A is located at the start of the genome map (bp 1); the remaining smaller annotations illustrate locations and directions of the fragmented rRNA coding regions (lsrDNA fragments – dark annotated regions; ssrDNA fragments – light annotated regions) interspersed amongst the three protein-coding genes.