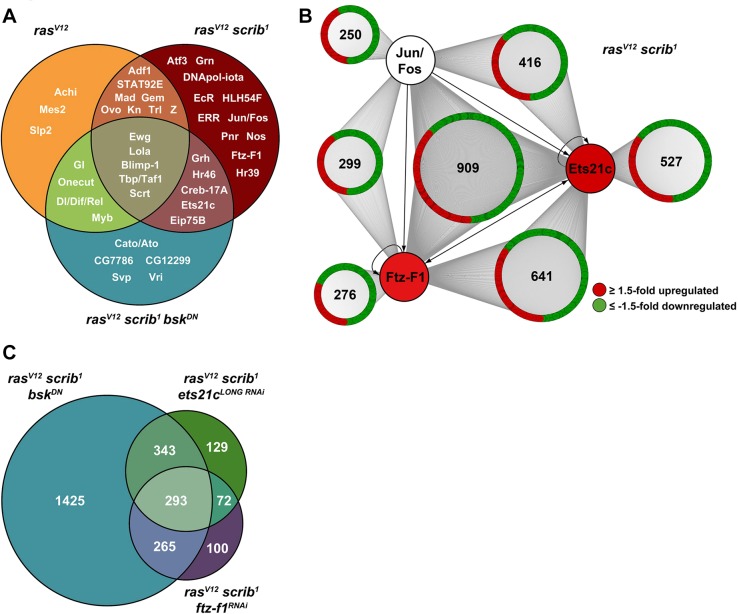
Fig. 2.
Transcription factor network orchestrates tumor-specific gene expression signature. (A) The number and diversity of enriched DNA motifs and hence putative TFs that regulate gene expression increase with tumor complexity as identified by iRegulon. Venn diagram shows specific enrichment of the binding sites for AP-1 factors (e.g. Jun/Fos), Atf3, NRs (e.g. Ftz-F1, EcR) in rasV12scrib1 mosaic EAD, whereas an Ets21c motif is overrepresented also among genes regulated in rasV12scrib1bskDN mosaic EAD. (B) Putative AP-1, Ftz-F1 and Ets21c binding motifs were found by FIMO 5 kb upstream and within first introns of numerous genes misregulated in rasV12scrib1 tumors. Of those genes, many contain binding sites for all three TFs or a combination of Ets21c/Ftz-F1 or Ets21c/AP-1 motifs. The network connects the candidate TFs to their putative target genes that are up- (red) or downregulated (green; ≥1.5-fold) in rasV12scrib1 tumors. In contrast to Jun and Fos, Ftz-F1 and Ets21c are themselves transcriptionally upregulated in rasV12scrib1 malignant tumors, possibly through a self-regulatory and/or AP-1-dependent mechanism (arrows). (C) Venn diagram shows intersection of genes that are misregulated in rasV12scrib1 tumors but rescued upon inhibition of JNK (rasV12scrib1bskDN), knockdown of ets21c (rasV12scrib1ets21cLONG RNAi) or ftz-f1 (rasV12scrib1ftz-f1RNAi). Rescue was defined as ≥1.5-fold change in expression from rasV12scrib1 towards control levels. (B,C) See supplementary material Table S1 for corresponding gene lists.