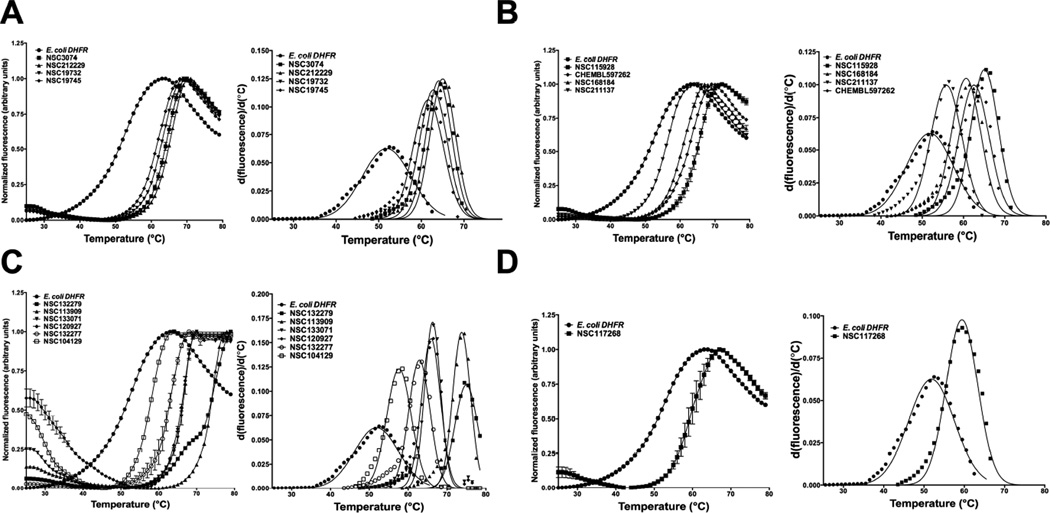
Fig. 2.
Differential scanning fluorimetry, DSF, curves and their first derivatives for E. coli DHFR in the presence of 15 analogues of 1,3,5-triazine-2,4-diamine. (A) DSF curves for halide, methyl, methoxy and ethoxy substituents at the R1 position of 1-phenyl-6,6-dimethyl-1,3,5-triazine-2,4-diamine. (B) DSF curves for nitrile, dimethylamino and aminomethyl substituents at either R1 or R2 position of 1-phenyl-6,6-dimethyl-1,3,5-triazine-2,4-diamine. (C) DSF curves for alkyl benzenesulfonyl fluoride, phenoxypropoxyphenyl, phenylbutyl, 2,4-dichlorophenylbutyl and fluorosulfonylphenylaminocarbonyl substituents at either R1 or R2 position of 1-phenyl-6,6-dimethyl-1,3,5-triazine-2,4-diamine. Note that this class of molecules showed the largest thermal shifts. (D) DSF curve for 4-chlorophenyl and sulfonamide substitution at the R1 and R3 position of 1-phenyl-1,3,5-triazine-4,6-diamine. The y-axis in primary unfolding curves represents normalized fluorescence and the x-axis shows the temperature in degrees Celsius. The inhibitors were kept fixed at 500 µM except NSC132277 and NSC132299, which were done at 10 µM concentration. Right panel in each plot shows the Gaussian fit of first-derivative for curves from left panel. Note that the molecules NSC117268, NSC133071, NSC168184, NSC104129, CHEMBL597262 and NSC333873 were also independently picked by the virtual ligand screening algorithm, PoLi.