FIG 4 .
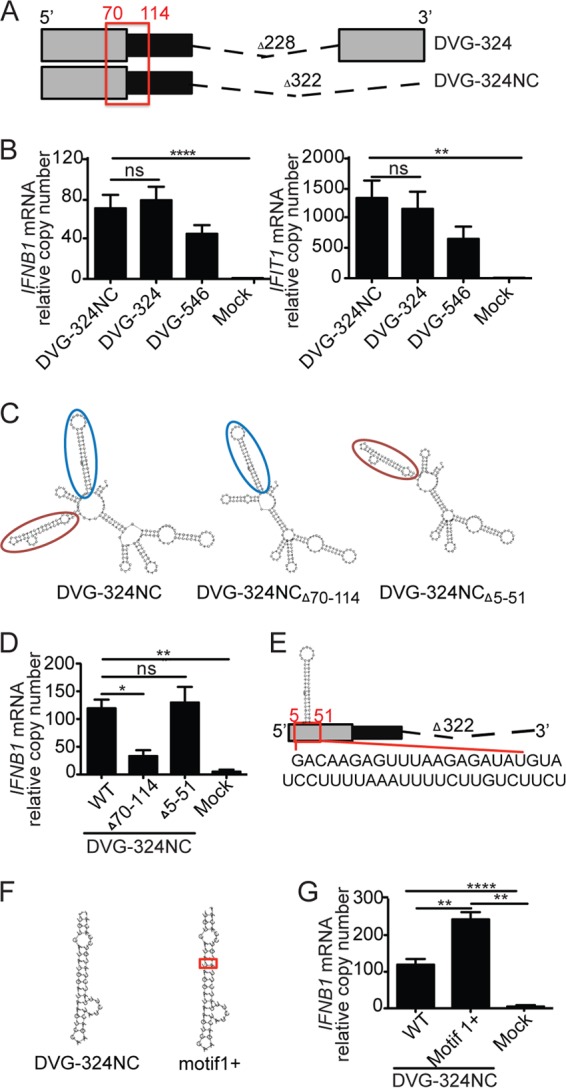
3′-to-5′ complementarity and additional proximal secondary structures are not necessary for IFN induction by DVG-derived RNA. (A) Illustration of DVG-324 and the related 3′ complementary sequence deletion mutant (DVG-324NC). The relative position of the immunostimulatory motif DVG70-114 is highlighted with a red square. The length of the deletion (dashed line) is indicated. (B) Expression of IFNB1 and IFIT1 mRNA by RT-qPCR of LLC-MK2 cells transfected for 6 h with 4.15 pmol of the ivtDVG indicated. (C) Folding predictions for DVG-324NCΔ70-114, DVG-324NCΔ5-51, and parental DVG-324NC. Motif DVG70-114 is indicated by a red oval. Motif DVG5-51 is indicated by a blue oval. (D) Expression of IFNB1 mRNA by RT-qPCR in LLC-MK2 cells transfected for 6 h with 4.15 pmol of the ivtDVG indicated. (E) Illustration of the DVG5-51 motif and its relative position in the DVG-324NC construct. The sequence of DVG5-51 is shown below the construct. (F) Folding prediction for DVG-324NC motif1+ compared to that for DVG-324NC. (G) Expression of IFNB1 mRNA by RT-qPCR of LLC-MK2 cells transfected for 6 h with 4.15 pmol of the ivtDVG illustrated in panel H. All transfection experiments were independently repeated at least three times. The data are the mean ± the standard error of the mean of all of the experiments (total n = 3 to 5/group). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant (one-way ANOVA with Bonferroni’s post hoc test). Data are expressed as the copy number relative to that of the housekeeping gene for GAPDH.
