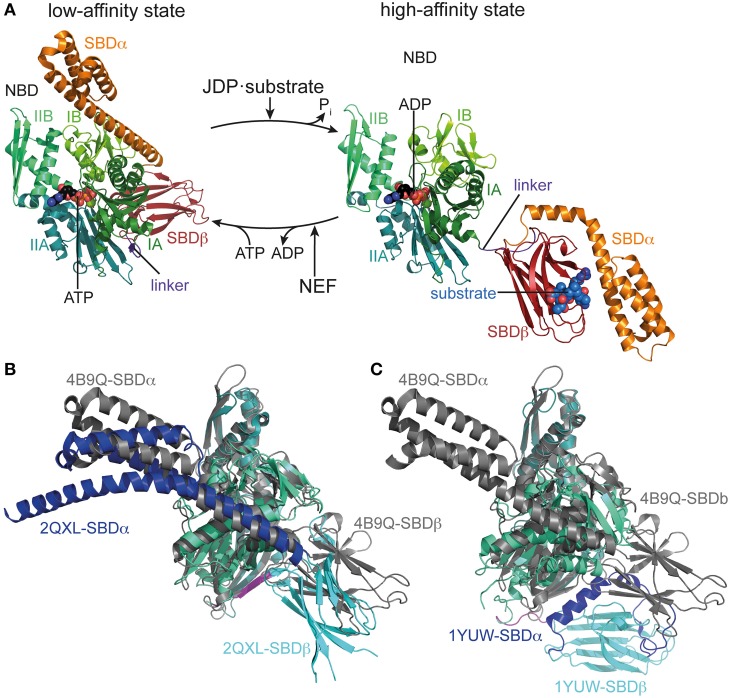
Figure 1.
Conformational cycle of Hsp70s. (A) Structural changes associated with the ATPase cycle of E. coli DnaK. Left, crystal structure of DnaK in the ATP bound open conformation (low-affinity state, PDB ID 4B9Q, Kityk et al., 2012) in cartoon representation with NBD subdomains IA, IB, IIA, IIB in different shades of green, SBDβ in dark red and SBDα in orange. Right, solution structure of DnaK in the ADP bound/nucleotide-free state as derived from residual dipolar coupling NMR experiments and crystal structures of the isolated domains (high-affinity state, PDB IDs 2KHO, Bertelsen et al., 2009) colored as in the ATP bound state and NBDs in identical orientation. (B) Overlay of the crystal structures of DnaK·ATP (PDB ID 4B9Q; gray) and Sse1 (PDB ID 2QXL, Liu and Hendrickson, 2007); NBD, deep teal; SBDβ, cyan; SBDα, blue. (C) Overlay of the crystal structures of DnaK·ATP (PDB ID 4B9Q; gray) and bovine Hsc70(1-554) (PDB ID 1YUW, Jiang et al., 2005) NBD, deep teal; SBDβ, cyan; SBDα, blue.