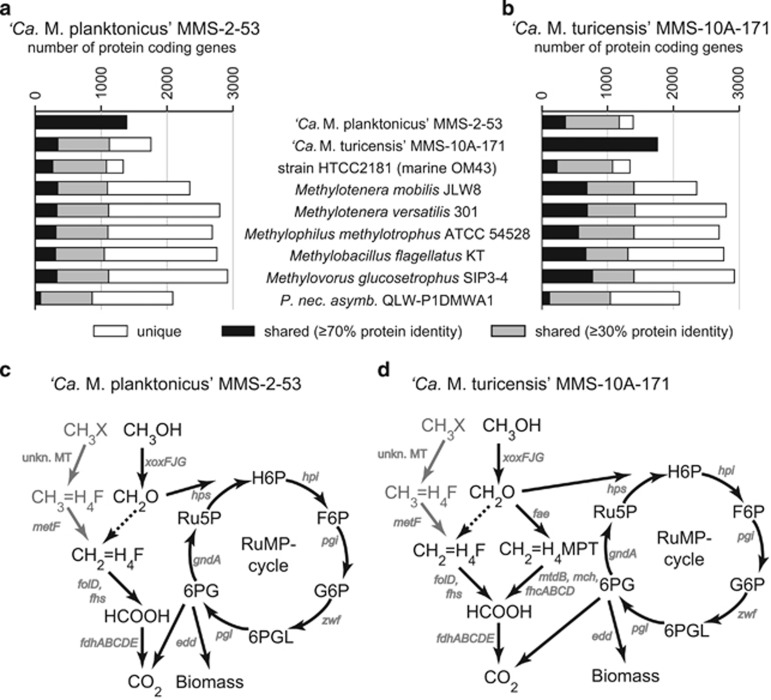
Figure 6.
(a, b) Number of protein-coding genes shared between ‘Ca. Methylopumilus planktonicus' MMS-2-53 (a) or ‘Ca. Methylopumilus turicensis' MMS-10A-171 (b) and genome sequenced members of Methylophilaceae and a typical freshwater microbe (Polynucleobacter necessarius subsp. asymbioticus QLW-P1DMWA-1). Protein identity cutoffs of ⩾30% and ⩾70% (minLrap⩾0.8) were applied. (c, d) Reconstruction of methylotrophic pathways of ‘Ca. Methylopumilus planktonicus' MMS-2-53 (c) and ‘Ca. Methylopumilus turicensis' MMS-10A-171 (d). Genes involved in methylotrophy are given in gray (see Supplementary Table S2 for more information). Dashed lines indicate spontaneous reactions, gray lines indicate possible pathways for CH3X oxidation. unkn. MT, unknown methyltransferase; CH3OH, methanol; CH3X, other C1 compounds, for example, methylamine; CH2O, formaldehyde; HCOOH, formate; H4F, tetrahydrofolate; H4MPT, tetrahydromethanopterin; H6P; hexulose-6-phosphate; F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phospho-gluconate; Ru5P, ribulose-5-phosphate; RuMP cycle, ribulose monophosphate cycle.