Fig. 1.
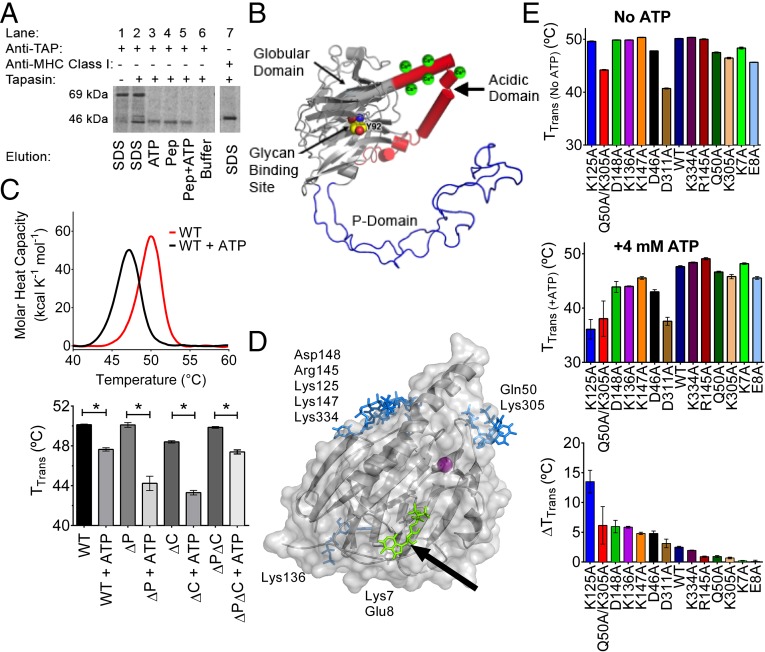
ATP induces dissociation of MHC class I from the PLC (A) and interacts with the globular domain of calreticulin (B–E). (A) Gel panel shows a representative anti-TAP IP of lysates from [35S]-methionine–labeled CRT−/− MEFs expressing mCRT(WT). The resulting IPs were eluted with SDS (lanes 1 and 2), ATP (lane 3), the SIINFEKL peptide (Pep; lane 4), the SIINFEKL peptide together with ATP (lane 5), or buffer alone (lane 6). Lane 1 is a negative control of an anti-TAP IP using lysates from tapasin-deficient cells that are impaired for MHC class I-TAP binding. Lane 7 shows a direct MHC class I (Y3) IP from CRT−/− MEFs to mark the migration position of MHC class I. Data are representative of three independent experiments. (B) Model for calreticulin showing its globular, P-terminal, and C-terminal domains. The globular domain structure (gray) is based on PDB ID code 3O0V (15), the P-domain structure (blue) is based on PDB ID code 1HHN (45), and the P-domain orientation is modeled based on PDB ID code 3RG0 (16). The acidic domain [modeled de novo using I-TASSER (46)] is indicated in red, with multiple low-affinity calcium-binding sites (shown as green spheres). The glycan-binding residue Tyr92 on the concave surface of the globular domain is indicated (atoms are represented as spheres with carbon colored yellow, oxygen colored red, and nitrogen colored blue). (C, Upper) Representative DSC thermograms showing the thermostability of calreticulin in the absence or presence of 4 mM ATP. (C, Lower) Quantification of thermostability change (TTrans) values for calreticulin constructs containing or lacking the P-domain, acidic domain, or both [mCRT(ΔP), mCRT(ΔC), and mCRT(ΔPΔC)] in the presence or absence of 4 mM ATP. Data show the mean ± SEM from two to three [mCRT(ΔC), mCRT(ΔP), and mCRT(ΔPΔC)] or 27 [mCRT(WT)] independent experiments. Statistically significant differences in the mean TTrans values (assessed via a one-way ANOVA, followed by a Tukey’s post hoc test) are denoted (*). (D) Global docking of ATP to the globular domain of mCRT [PDB ID code 3O0V (15)]. Key nucleotide-interacting residues from each cluster are noted. An arrow denotes the cluster of poses in close proximity to Lys7 (green). (E) Interaction of calreticulin point mutants with ATP. Plots show the mean TTrans values (±SEM) in the absence of ATP (Upper) and in the presence of 4 mM ATP (Middle), and the ATP-induced change in the TTrans values (ΔTTrans) (Lower). Data represent the mean TTrans ± SEM from two to four (mutants) or 27 [mCRT(WT)] independent experiments.