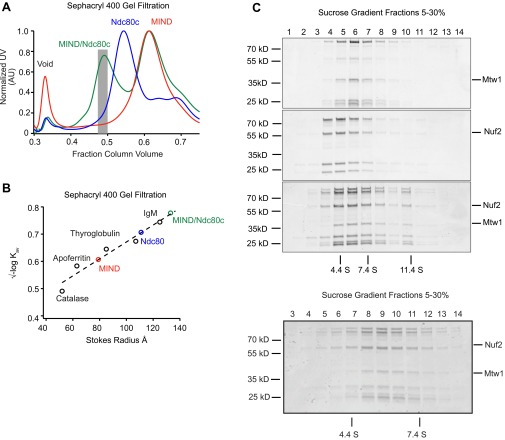
Fig. S1.
MIND/Ndc80c co-complex associates with 1:1 stoichiometry. (A) Representative Sephacryl 400 gel filtration elution profiles for Ndc80c (green), MIND (blue), and MIND/Ndc80c co-complex (red). Note that, for the MIND/Ndc80c experiment, complexes were mixed in a 2.5:1 molar ratio (MIND:Ndc80c); therefore, excess MIND can be seen eluting as a separate peak. Gray bar indicates fractions pooled and used for subsequent MIND/Ndc80c experiments. (B) Stokes radius vs. √–logKav plotted for indicated standard proteins (catalase, 52 Å; apoferritin, 63 Å; thyroglobulin, 85 Å; fibrinogen, 107 Å; IgM, 130 Å) and fit with a linear regression. Stokes radii for Ndc80c, MIND, and MIND/Ndc80c were calculated using the equation derived from the linear fit: Ndc80c = 111 Å, MIND = 79 Å, MIND/Ndc80c = 134 Å. (C) Velocity sedimentation analysis of Ndc80c, MIND, and MIND/Ndc80c run on a 5–30% sucrose gradient split into 16 sequential fractions; n = 2. Representative elution fractions for each complex across gradient are shown in Coomassie-stained gel; fraction 1 corresponds to the top of the gradient. Some stoichiometric MIND/Ndc80c oligomers are visible in later fractions. BSA (4.4 S), aldolase (7.4 S), and catalase (11.4 S) were run as standards (positions indicated by vertical lines), and a linear fit was determined to find the S values for experimental complexes: Ndc80c = 4.24 S, MIND = 5.42 S, MIND/Ndc80c = 5.89 S. Using the S value and stokes radius, the molecular mass of the MIND/Ndc80c complex was determined to be 331.8 kDa, within 0.04% of the expected molecular mass of a stoichiometric MIND/Ndc80c complex (333 kDa). The frictional ratio of MIND/Ndc80c was also calculated using the S value and stokes radius. MIND/Ndc80c exhibits a frictional ratio of 2.7, indicating that it is highly elongated. (Below) A sucrose gradient from a repeat experiment split into 18 sequential fractions to more clearly visualize the MIND/Ndc80c peak centered at 5.89 S.