Fig. 1.
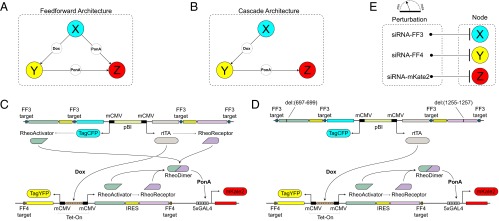
The benchmark synthetic regulatory networks. (A) The first motif is a coherent feed-forward loop where node X regulates node Z in both a direct and an indirect manner. (B) The second motif is a cascade, where node X activates node Z only by activating node Y. (C and D) Detailed information about the synthetic gene networks. The activity of the three nodes X, Y, and Z can be quantified by the output fluorescent proteins TagCFP, TagYFP and mKate2, respectively. The constitutive bidirectional promoter of node X also transcribes rtTA for node Y induction and the RheoSwitch dimers for node Z induction. For the cascade motif, the translation of RheoSwitch dimer protein is prevented by nonsense mutation. In the presence of doxycycline, the constitutively transcribed rtTA induces transcription of RheoSwitch in node Y. When ponasterone A binds to the RheoSwitch dimer, the entire complex serves as a transactivator for the yeast Gal4 domain. Transcription at Gal4 domain results in production of mKate2 to indicate node Z activity. (E) Perturbation of each node in the system is performed siRNA. Nodes X and Y are perturbed by synthetic siRNAs (FF3 and FF4, respectively) with the targets located in the 3′ UTR of their corresponding targets. Node Z is perturbed by a custom siRNA that directly targets mKate2. IRES, internal ribosome entry site.
