Fig. S3.
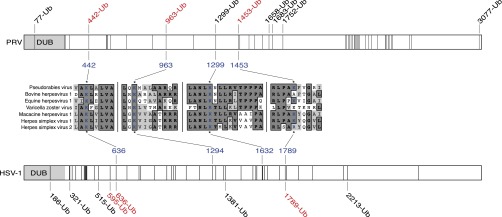
Local alignments of pUL36 lysines from seven neuroinvasive herpesviruses annotated with results from UbiScan mass spectrometry analysis of PRV and HSV-1 pUL36. Illustration of pUL36 lysine residues in PRV (Top, 49 lysines) and HSV-1 (Bottom, 48 lysines). Lysines are represented as vertical lines, with ubiquitinated lysines (Ub) indicated by amino acid position. Lysines that are DUB autocatalytic substrate sites are indicated in red. Local sequence alignments for the three lysines studied (PRV K442, K963, and K1299) are shown with corresponding HSV-1 lysines indicated. An alignment for a fourth lysine that is a DUB substrate in PRV and HSV-1, PRV K1453, is also provided. Alignment details: conserved amino acids are in bold with dark gray background, and amino acids with similar properties have a light gray background.
