Figure 4. Changes in DNA Methylation Patterns in Adaptive NK Cells Are Similar to Those in Cytotoxic T Cells.
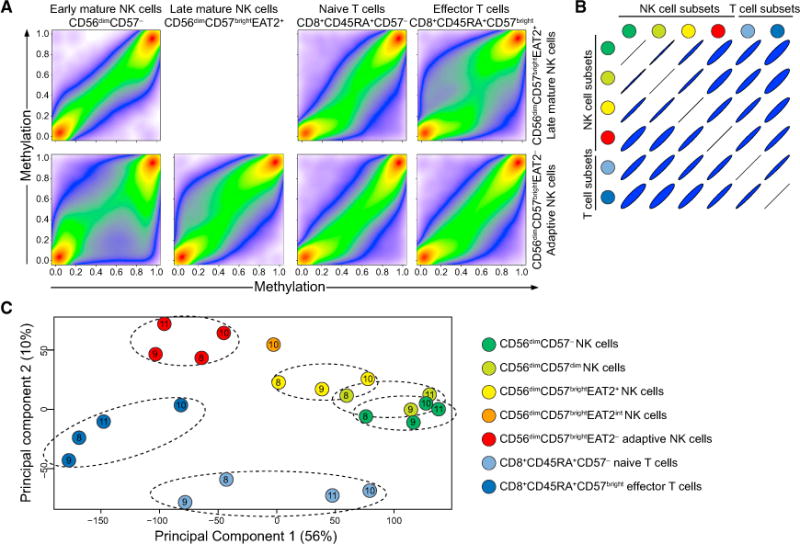
(A–C) NK and T cell subsets were sorted from four HCMV+ donors. Genomic DNA from each subset was treated with sodium bisulfite and subjected to genome-wide DNA methylation profiling.
(A) Pseudo-color scatterplots illustrating multiple pairwise comparisons of methylation (β-value) over all probes from specific subsets in donor 10 after data filtering and probe normalization. A β-value close to zero corresponds with no methylation, and a β-value close to 1 corresponds with full methylation.
(B) Correlation matrix of all pairwise subset comparisons.
(C) MDS analysis of DNA methylation in NK cell and T cell subsets after data filtering and normalization. Samples are color coded based on the cell type and labeled according to the donor they were derived from (donors 8, 9, 10, and 11). Only probes with a high variability in β-values across the different cell subsets are shown (arbitrarily set as a ratio of standard deviation over a mean greater than 0.1, n = 271,769). Dimensions 1 and 2 represent the largest source of variation (totaling 66%).
See also Figure S4.
